You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003452_01194
You are here: Home > Sequence: MGYG000003452_01194
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
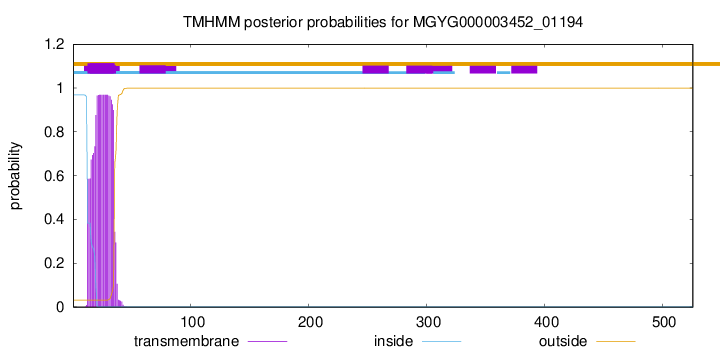
TMHMM annotations
Basic Information help
| Species | Bifidobacterium ruminantium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium ruminantium | |||||||||||
| CAZyme ID | MGYG000003452_01194 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7470; End: 9050 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3583 | YabE | 2.94e-41 | 16 | 282 | 15 | 282 | Uncharacterized conserved protein YabE, contains G5 and tandem DUF348 domains [Function unknown]. |
| pfam07501 | G5 | 9.41e-21 | 212 | 284 | 3 | 75 | G5 domain. This domain is found in a wide range of extracellular proteins. It is found tandemly repeated in up to 8 copies. It is found in the N-terminus of peptidases belonging to the M26 family which cleave human IgA. The domain is also found in proteins involved in metabolism of bacterial cell walls suggesting this domain may have an adhesive function. |
| pfam03990 | DUF348 | 1.40e-07 | 41 | 77 | 1 | 37 | Domain of unknown function (DUF348). This domain normally occurs as tandem repeats; however it is found as a single copy in the S. cerevisiae DNA-binding nuclear protein YCR593. This protein is involved in sporulation part of the SET3C complex, which is required to repress early/middle sporulation genes during meiosis. The bacterial proteins are likely to be involved in a cell wall function as they are found in conjunction with the pfam07501 domain, which is involved in various cell surface processes. |
| NF035936 | agg_sub_LPXTH | 1.05e-05 | 310 | 394 | 52 | 136 | serine-rich aggregation substance UasX. Members of this protein family are repetitive, serine-rich surface proteins of the Firmicutes, found primarily in the genus Leuconostoc. The variant form of sortase signal, LPXTH, is replaced by LPXTG in members from some lineages, such as Weissella oryzae, and therefore recognizable. Some members of this family have the KxYKxGKxW type signal peptide as seen in the glycoprotein adhesin GspB, a substrate of the accessory Sec system for secretion. WOSG25_050600 from Weissella oryzae SG25 is identified in a publication as an unnamed aggregation substance, a conclusion supported by the sorting signals and composition reported here. We assign the gene symbol uasX (unnamed aggregation substance X) based on our evaluation of the family. |
| NF035936 | agg_sub_LPXTH | 1.21e-05 | 307 | 395 | 24 | 113 | serine-rich aggregation substance UasX. Members of this protein family are repetitive, serine-rich surface proteins of the Firmicutes, found primarily in the genus Leuconostoc. The variant form of sortase signal, LPXTH, is replaced by LPXTG in members from some lineages, such as Weissella oryzae, and therefore recognizable. Some members of this family have the KxYKxGKxW type signal peptide as seen in the glycoprotein adhesin GspB, a substrate of the accessory Sec system for secretion. WOSG25_050600 from Weissella oryzae SG25 is identified in a publication as an unnamed aggregation substance, a conclusion supported by the sorting signals and composition reported here. We assign the gene symbol uasX (unnamed aggregation substance X) based on our evaluation of the family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAF40395.1 | 1.31e-244 | 1 | 526 | 1 | 506 |
| AVT46031.1 | 2.00e-244 | 1 | 526 | 1 | 508 |
| AXR42445.1 | 2.59e-242 | 1 | 526 | 1 | 507 |
| QHB63546.1 | 2.59e-242 | 1 | 526 | 1 | 507 |
| AII77134.1 | 3.68e-242 | 1 | 526 | 1 | 507 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6M6N7 | 1.42e-13 | 38 | 268 | 42 | 273 | Resuscitation-promoting factor Rpf2 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=rpf2 PE=1 SV=1 |
| P37546 | 3.02e-11 | 38 | 287 | 37 | 288 | Putative cell wall shaping protein YabE OS=Bacillus subtilis (strain 168) OX=224308 GN=yabE PE=2 SV=2 |
| A0R3E0 | 1.25e-06 | 27 | 282 | 13 | 269 | Resuscitation-promoting factor RpfB OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=rpfB PE=3 SV=2 |
| Q8CQD9 | 3.22e-06 | 214 | 286 | 1223 | 1297 | Accumulation-associated protein OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=SE_0175 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
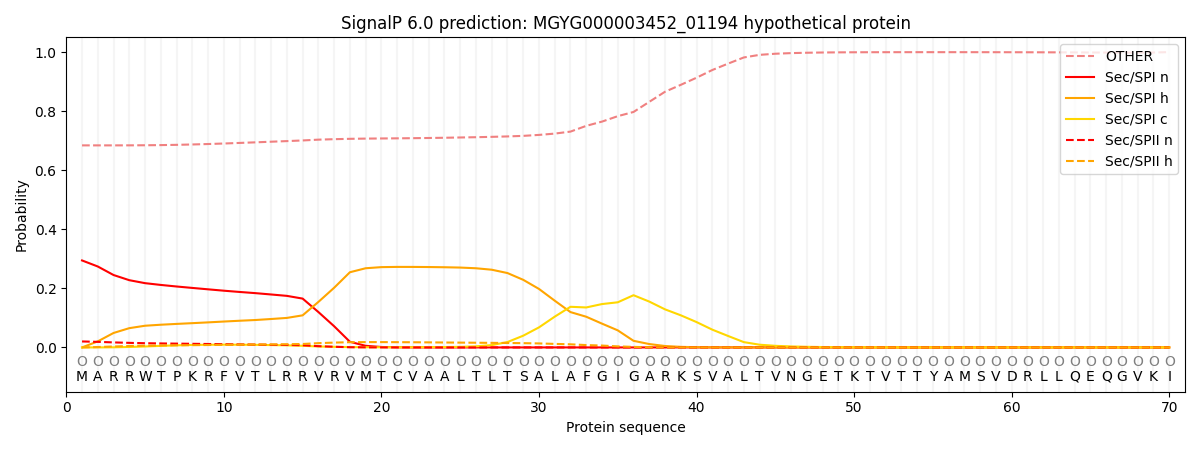
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.691575 | 0.284967 | 0.021265 | 0.000473 | 0.000248 | 0.001477 |