You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003479_00125
You are here: Home > Sequence: MGYG000003479_00125
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900548275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900548275 | |||||||||||
| CAZyme ID | MGYG000003479_00125 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41101; End: 42207 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 48 | 321 | 3.6e-109 | 0.9963636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.32e-05 | 162 | 312 | 106 | 266 | Cellulase (glycosyl hydrolase family 5). |
| pfam00331 | Glyco_hydro_10 | 6.03e-05 | 97 | 227 | 58 | 162 | Glycosyl hydrolase family 10. |
| COG3934 | COG3934 | 6.88e-05 | 37 | 188 | 4 | 148 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| COG3693 | XynA | 8.51e-05 | 7 | 227 | 6 | 186 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLC65017.1 | 6.34e-125 | 14 | 364 | 34 | 383 |
| AOC97220.1 | 3.62e-124 | 15 | 364 | 35 | 383 |
| ABQ05053.1 | 5.70e-123 | 20 | 364 | 40 | 383 |
| QDW21585.1 | 8.34e-123 | 15 | 364 | 35 | 383 |
| AWK07461.1 | 9.25e-122 | 20 | 364 | 40 | 383 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
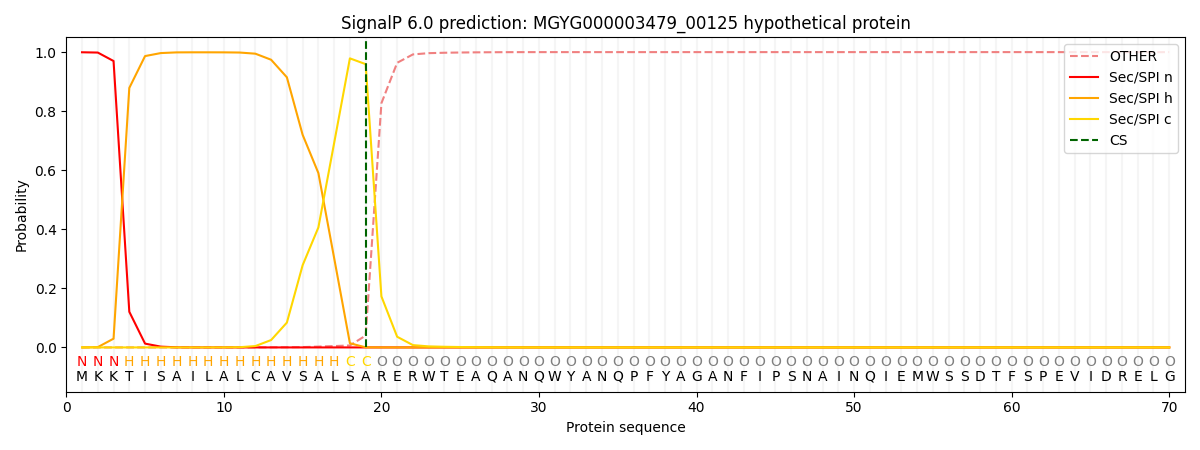
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000541 | 0.997427 | 0.001347 | 0.000240 | 0.000220 | 0.000199 |
