You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003482_01367
You are here: Home > Sequence: MGYG000003482_01367
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Selenomonas_C bovis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Selenomonadales; Selenomonadaceae; Selenomonas_C; Selenomonas_C bovis | |||||||||||
| CAZyme ID | MGYG000003482_01367 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | Mannan endo-1,4-beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 117884; End: 119224 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 67 | 373 | 7.5e-61 | 0.8382838283828383 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 3.25e-43 | 67 | 394 | 1 | 286 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 3.40e-19 | 207 | 378 | 145 | 324 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAL83629.1 | 3.10e-184 | 1 | 441 | 1 | 444 |
| AME03684.1 | 1.68e-165 | 8 | 445 | 14 | 453 |
| AKT53686.1 | 6.81e-165 | 23 | 445 | 29 | 453 |
| AOH46966.1 | 3.16e-163 | 1 | 445 | 10 | 453 |
| BBK75947.1 | 1.03e-131 | 61 | 443 | 289 | 675 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 4.85e-57 | 66 | 429 | 51 | 414 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 2BVT_A | 4.37e-52 | 60 | 444 | 2 | 372 | Thestructure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVT_B The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVY_A The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi [Cellulomonas fimi] |
| 2X2Y_A | 3.25e-50 | 60 | 444 | 2 | 372 | Cellulomonasfimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi],2X2Y_B Cellulomonas fimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi] |
| 1J9Y_A | 4.77e-50 | 61 | 392 | 6 | 324 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1R7O_A | 6.02e-50 | 61 | 392 | 16 | 334 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 6.18e-49 | 61 | 392 | 44 | 362 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 9.18e-48 | 35 | 446 | 9 | 412 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P16699 | 5.35e-19 | 164 | 321 | 123 | 273 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| O05512 | 1.17e-18 | 163 | 318 | 121 | 268 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 5.16e-18 | 209 | 318 | 158 | 266 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
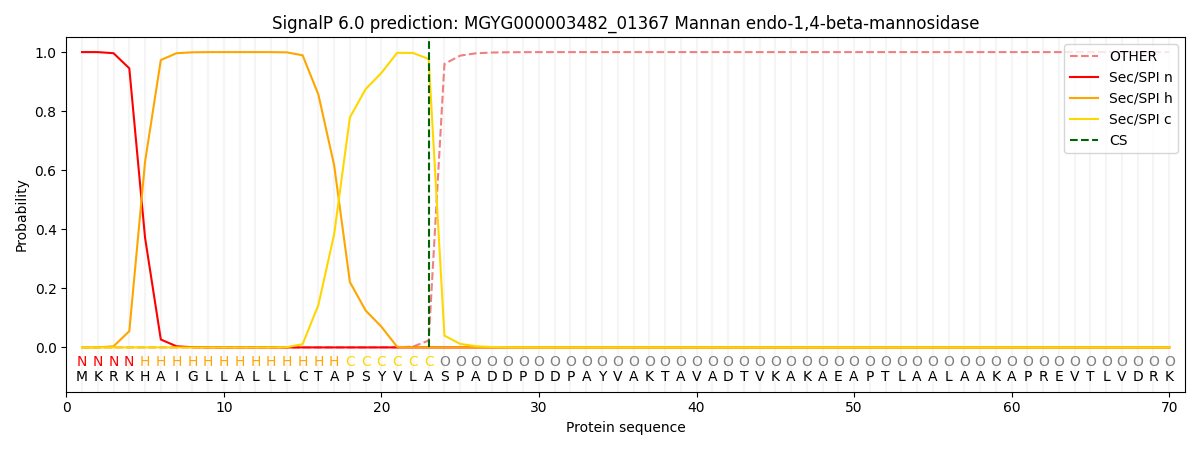
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000278 | 0.999048 | 0.000202 | 0.000176 | 0.000155 | 0.000143 |
