You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003482_02066
You are here: Home > Sequence: MGYG000003482_02066
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Selenomonas_C bovis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Selenomonadales; Selenomonadaceae; Selenomonas_C; Selenomonas_C bovis | |||||||||||
| CAZyme ID | MGYG000003482_02066 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48520; End: 49380 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 156 | 281 | 1.7e-16 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01832 | Glucosaminidase | 6.06e-12 | 157 | 221 | 3 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| NF033838 | PspC_subgroup_1 | 2.94e-04 | 37 | 114 | 361 | 434 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNH54403.1 | 4.54e-104 | 104 | 286 | 76 | 258 |
| AOH47456.1 | 4.58e-103 | 87 | 286 | 55 | 254 |
| AKT54187.1 | 8.32e-103 | 87 | 286 | 53 | 251 |
| AME03076.1 | 4.78e-102 | 87 | 286 | 53 | 251 |
| AEB99772.1 | 3.02e-99 | 13 | 286 | 13 | 266 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 3.42e-20 | 132 | 266 | 50 | 179 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
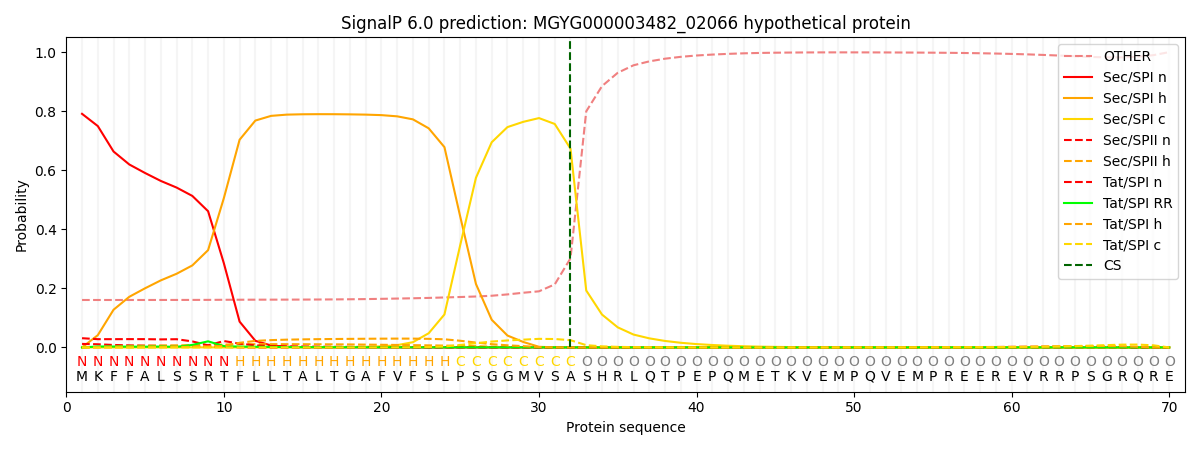
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.186567 | 0.753605 | 0.013702 | 0.037320 | 0.007381 | 0.001393 |
