You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003483_00917
You are here: Home > Sequence: MGYG000003483_00917
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
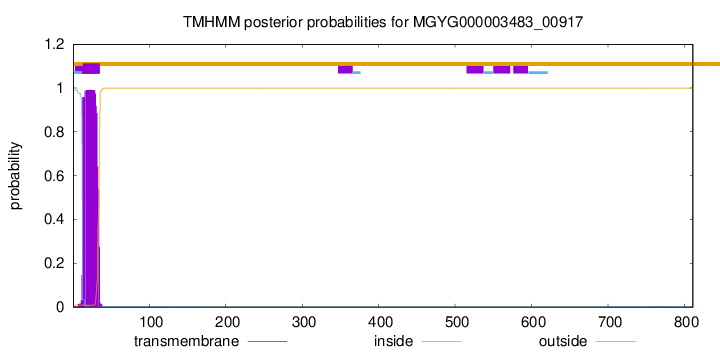
TMHMM annotations
Basic Information help
| Species | RUG572 sp900547945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900547945 | |||||||||||
| CAZyme ID | MGYG000003483_00917 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4033; End: 6468 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 31 | 720 | 2.2e-82 | 0.6223404255319149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.99e-56 | 33 | 754 | 14 | 682 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 4.86e-13 | 104 | 475 | 112 | 451 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 8.43e-08 | 299 | 475 | 239 | 419 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 5.84e-07 | 257 | 340 | 21 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 9.92e-07 | 33 | 190 | 3 | 157 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23305.1 | 1.07e-245 | 23 | 802 | 20 | 796 |
| AGB28636.1 | 2.40e-234 | 32 | 802 | 27 | 791 |
| QNL40264.1 | 9.94e-234 | 16 | 802 | 9 | 791 |
| QQR15691.1 | 5.64e-233 | 16 | 802 | 9 | 791 |
| QRM99259.1 | 5.64e-233 | 16 | 802 | 9 | 791 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5N6U_A | 3.00e-91 | 32 | 761 | 27 | 713 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6BYE_A | 4.47e-78 | 30 | 755 | 4 | 723 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYC_A | 4.54e-78 | 30 | 755 | 4 | 723 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 2.97e-77 | 30 | 755 | 2 | 721 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 3.02e-77 | 30 | 755 | 4 | 723 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0CCA0 | 2.37e-71 | 67 | 733 | 30 | 677 | Beta-mannosidase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=mndB PE=3 SV=2 |
| Q29444 | 8.41e-69 | 14 | 702 | 4 | 684 | Beta-mannosidase OS=Bos taurus OX=9913 GN=MANBA PE=1 SV=1 |
| A1CGA8 | 1.72e-67 | 69 | 727 | 32 | 673 | Beta-mannosidase B OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=mndB PE=3 SV=1 |
| I2C092 | 6.89e-67 | 32 | 704 | 7 | 654 | Beta-mannosidase B OS=Thermothelomyces thermophilus OX=78579 GN=man9 PE=1 SV=1 |
| Q95327 | 8.31e-66 | 14 | 702 | 4 | 684 | Beta-mannosidase OS=Capra hircus OX=9925 GN=MANBA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
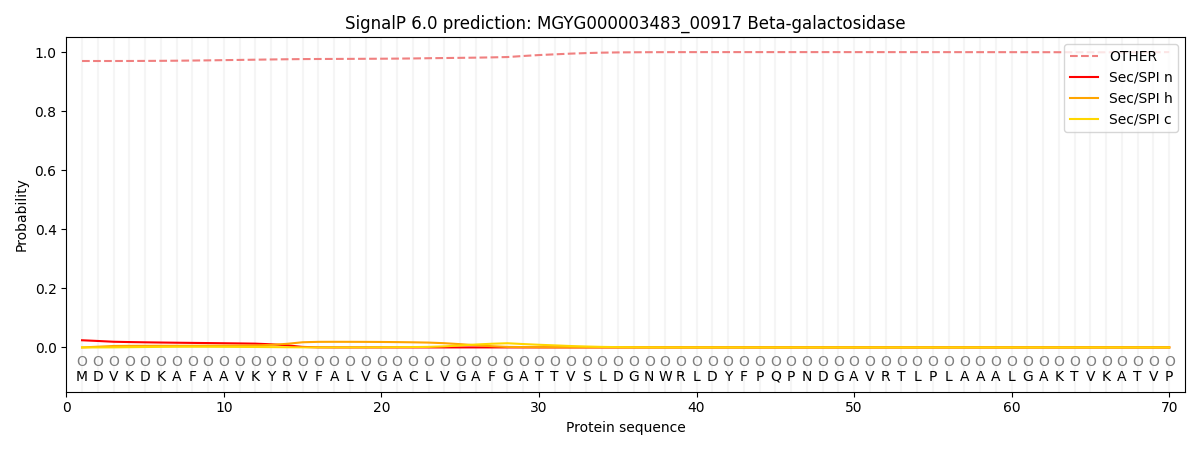
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.971208 | 0.022111 | 0.004830 | 0.000133 | 0.000084 | 0.001658 |