You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003483_01946
Basic Information
help
| Species |
RUG572 sp900547945
|
| Lineage |
Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900547945
|
| CAZyme ID |
MGYG000003483_01946
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 677 |
|
75453.29 |
8.2366 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003483 |
3685575 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 8655;
End: 10688
Strand: -
|
No EC number prediction in MGYG000003483_01946.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
133 |
359 |
6.4e-33 |
0.8021978021978022 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG3934
|
COG3934 |
3.07e-12 |
50 |
386 |
5 |
308 |
Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
A1DBV1
|
4.02e-06 |
110 |
321 |
168 |
392 |
Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manF PE=3 SV=1 |
This protein is predicted as SP
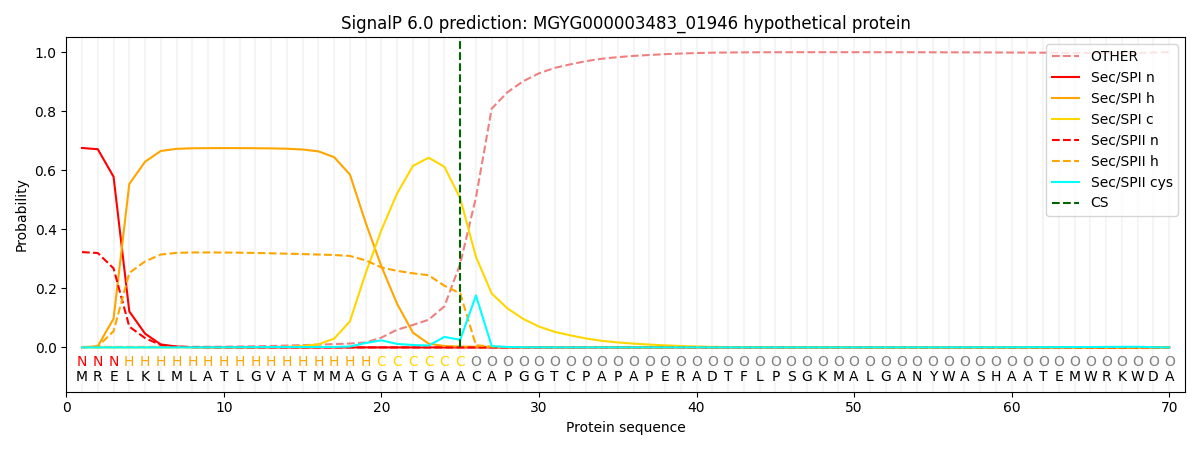
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.004440
|
0.660757
|
0.333505
|
0.000606
|
0.000376
|
0.000300
|
There is no transmembrane helices in MGYG000003483_01946.

