You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003483_02131
You are here: Home > Sequence: MGYG000003483_02131
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RUG572 sp900547945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900547945 | |||||||||||
| CAZyme ID | MGYG000003483_02131 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4513; End: 6447 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 57 | 334 | 7.3e-57 | 0.9927272727272727 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12146 | Hydrolase_4 | 5.40e-05 | 505 | 616 | 80 | 224 | Serine aminopeptidase, S33. This domain is found in bacteria and eukaryotes and is approximately 110 amino acids in length. It is found in association with pfam00561. The majority of the members in this family carry the exopeptidase active-site residues of Ser-122, Asp-239 and His-269 as in UniProtKB:Q7ZWC2. |
| COG0596 | MhpC | 5.48e-05 | 502 | 641 | 89 | 278 | Pimeloyl-ACP methyl ester carboxylesterase [Coenzyme transport and metabolism, General function prediction only]. |
| pfam00561 | Abhydrolase_1 | 5.92e-05 | 492 | 631 | 54 | 245 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| PRK14875 | PRK14875 | 7.95e-04 | 501 | 627 | 197 | 353 | acetoin dehydrogenase E2 subunit dihydrolipoyllysine-residue acetyltransferase; Provisional |
| COG2267 | PldB | 0.003 | 505 | 641 | 111 | 291 | Lysophospholipase, alpha-beta hydrolase superfamily [Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOV18692.1 | 7.81e-106 | 31 | 388 | 4 | 358 |
| AVM44569.1 | 5.16e-104 | 31 | 380 | 5 | 346 |
| AUS96929.1 | 1.23e-78 | 31 | 386 | 5 | 353 |
| AYQ73077.1 | 5.49e-62 | 31 | 381 | 7 | 358 |
| QNP56058.1 | 5.76e-61 | 28 | 389 | 2 | 365 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
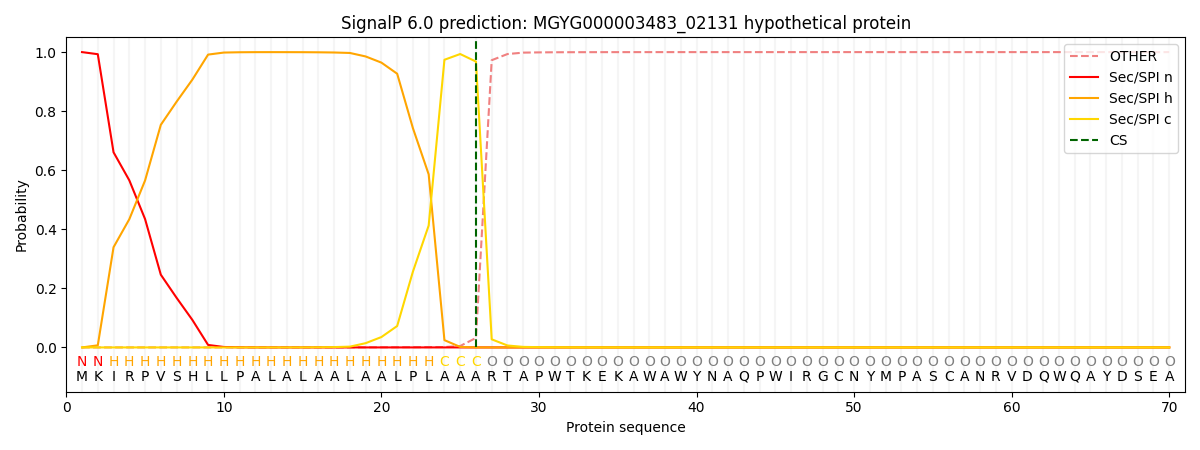
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000565 | 0.998648 | 0.000253 | 0.000188 | 0.000162 | 0.000155 |
