You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003493_00652
You are here: Home > Sequence: MGYG000003493_00652
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
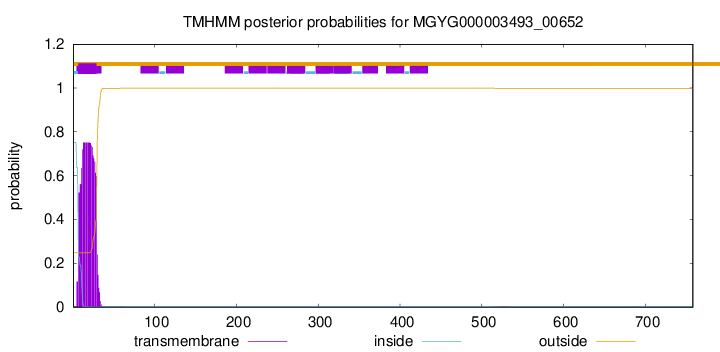
TMHMM annotations
Basic Information help
| Species | Prevotella sp002299275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002299275 | |||||||||||
| CAZyme ID | MGYG000003493_00652 | |||||||||||
| CAZy Family | CBM36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18005; End: 20281 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH8 | 215 | 529 | 1.1e-80 | 0.946875 |
| CBM36 | 34 | 147 | 1.4e-22 | 0.9652173913043478 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3405 | BcsZ | 7.33e-45 | 171 | 540 | 7 | 353 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam01270 | Glyco_hydro_8 | 4.64e-26 | 223 | 529 | 29 | 317 | Glycosyl hydrolases family 8. |
| cd04078 | CBM36_xylanase-like | 2.10e-24 | 30 | 148 | 4 | 119 | Carbohydrate Binding Module family 36 (CBM36); appended mainly to glycoside hydrolase family 11 (GH11) domains; xylan binding. This family includes carbohydrate binding module family 36 (CBM36) most of which appear appended to glycoside hydrolase family 11 (GH11) domains. These CBMs are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH11 catalytic modules with their dedicated, insoluble substrates. GH11 domains have xylanase (endo-1,4-beta-xylanase) activity which catalyzes the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. This family includes XynB from Dictyoglomus thermophilum Rt46B.1 and Xyn11A from Pseudobutyrivibrio xylanivorans Mz5T. Xyn11A is a multicatalytic enzyme with an N-terminal GH11 domain, a CBM36 domain, and a C-terminal putative NodB-like polysaccharide deacetylase which is predicted to be an acetyl esterase involved in debranching activity in the xylan backbone. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. Consistent with its structural and sequence similarity to CBM6, CBM36 binds xylan, but only at binding site I, and in a calcium-dependent manner; the latter suggests its potential application in affinity labeling. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUL54973.1 | 2.18e-168 | 36 | 541 | 41 | 543 |
| QWG04463.1 | 1.57e-153 | 28 | 538 | 48 | 563 |
| ANQ52232.1 | 2.08e-153 | 28 | 538 | 48 | 563 |
| ALQ07930.1 | 4.02e-141 | 41 | 540 | 39 | 552 |
| ASB49713.1 | 4.87e-137 | 47 | 536 | 184 | 679 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6G00_A | 2.17e-94 | 162 | 540 | 2 | 396 | CrystalStructure of a GH8 xylanase from Teredinibacter turnerae [Teredinibacter turnerae T7901],6G09_A Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae [Teredinibacter turnerae T7901],6G0B_A Crystal Structure of a GH8 xylotriose complex from Teredinibacter Turnerae [Teredinibacter turnerae T7901] |
| 6G0N_A | 1.18e-93 | 162 | 540 | 2 | 396 | CrystalStructure of a GH8 catalytic mutant xylohexaose complex xylanase from Teredinibacter turnerae [Teredinibacter turnerae T7901] |
| 5YXT_A | 7.32e-88 | 165 | 540 | 6 | 378 | Glycosidehydrolase family 8 Xylanase [Paenibacillus barengoltzii G22],5YXT_B Glycoside hydrolase family 8 Xylanase [Paenibacillus barengoltzii G22],5YXT_C Glycoside hydrolase family 8 Xylanase [Paenibacillus barengoltzii G22],5YXT_D Glycoside hydrolase family 8 Xylanase [Paenibacillus barengoltzii G22] |
| 6SRD_A | 8.66e-85 | 165 | 539 | 7 | 381 | Structureof Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis],6SRD_B Structure of Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis] |
| 6SUD_A | 8.66e-85 | 165 | 539 | 7 | 381 | Structureof L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis],6SUD_B Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A0S2UQQ5 | 6.25e-84 | 165 | 538 | 7 | 380 | Reducing-end xylose-releasing exo-oligoxylanase Rex8A OS=Paenibacillus barcinonensis OX=198119 GN=rex8A PE=1 SV=1 |
| Q9KB30 | 1.29e-80 | 165 | 540 | 7 | 381 | Reducing end xylose-releasing exo-oligoxylanase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=BH2105 PE=1 SV=1 |
| A1A048 | 5.39e-61 | 168 | 536 | 10 | 377 | Reducing end xylose-releasing exo-oligoxylanase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=xylA PE=1 SV=1 |
| P37701 | 3.90e-32 | 223 | 535 | 93 | 389 | Endoglucanase 2 OS=Ruminiclostridium josui OX=1499 GN=celB PE=3 SV=1 |
| P37699 | 1.73e-31 | 223 | 535 | 93 | 389 | Endoglucanase C OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCC PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
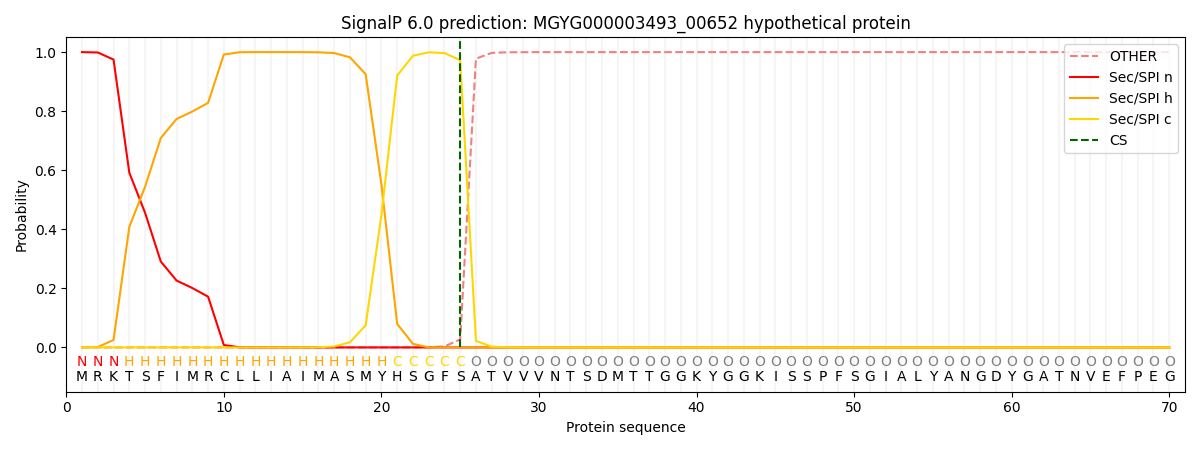
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000280 | 0.998944 | 0.000248 | 0.000173 | 0.000157 | 0.000147 |