You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003500_00112
You are here: Home > Sequence: MGYG000003500_00112
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
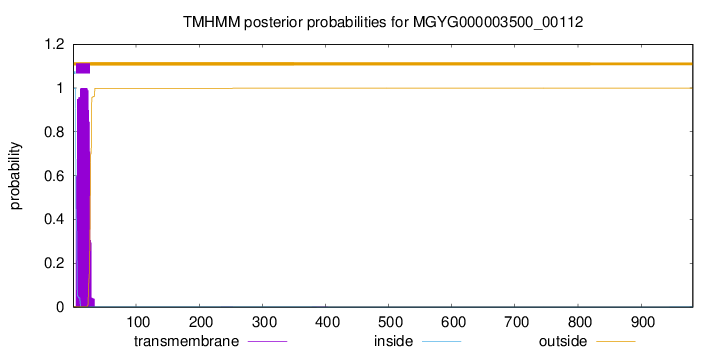
TMHMM annotations
Basic Information help
| Species | Butyrivibrio_A sp900768755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Butyrivibrio_A; Butyrivibrio_A sp900768755 | |||||||||||
| CAZyme ID | MGYG000003500_00112 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | Endoglucanase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 125493; End: 128441 Strand: - | |||||||||||
Full Sequence Download help
| MKKKVIITVA SILLGLALIV GTVVLACSVR QNIINDINHT SFEIEALGVI ESTKRNFDED | 60 |
| ALSGETTHDA TAGETSYEAS SEEICGDMTS EEDTQPEPSR EDSENASSGE HTQEAASPND | 120 |
| SGKATSGEKV TEKTSSGNDN RGENSTKEPA TIQNNDNSHN AVGDENGEGY EGIKGFGSYN | 180 |
| YGEALQKSIL FYELQRCGKL PETVRCNWRG DSCLNDGSDV GLDLTGGWID AGDNVKFNLP | 240 |
| MSYSAAMLAW SIYEDYDAYR ESGQLEYALA NIKWANDYFI KCHPEDEVYY YQVGNGGADH | 300 |
| GFWGPVEVVE YKMDRPSYKV TAQAPGSAVT GETAASLALC SILYKDIDRA YSELCLKHAK | 360 |
| SLYAFSEKYK SDAGYTAANG FYNSWSGFYD ELAWAGAWLY LATGDKTYIS KAKEFYPKGC | 420 |
| QDYNWAMCWD DVHIGAAVML SRITGDKVYS EAVEKHLDYW TTGTKDGQRI KYTPKGLAWL | 480 |
| DQWGSLRYAS TTAFIALVYT DDSACSASRK TAYVNFAEKQ ANYILGDTGF SYLIGFGDDY | 540 |
| PKNPHHRTAQ GSYADNMNEP SYSRHTLYGA LVGGPDASDN YSDKVSDYVC NEVACDYNAG | 600 |
| FTGLLAKMYS KYRGETIANF GAIETIDEKE YYVEAGINAS GTDFMEIKAI VYNKTAWPAR | 660 |
| ASKNAELRYF IDLSEVYDAG SNASGITIST NYMQDAFADG LKVWDEDNHI YYLSVRFLDG | 720 |
| KLYPGGQDKY KKEIQVRMKS SCGVWNNDND PSFAGMRTGS LSDGIRLALY EDGKLVYGTE | 780 |
| PPKGIDTGNT VIGGGTITSG SNVDSGGNSS GNNEDNSGNT ETQPTETAGN SSSESAQSGD | 840 |
| LSVNINYSDS SVGGIMTVKN NGSGTIQLSD LEIKYYFTKD NDTALQFACY HSAVQTAEGA | 900 |
| YNAVSGCEGR FVDYGLTDAD TLCTISFSDS TTLPGGAVLT VNFNINHSDW SAFNLSNDYS | 960 |
| KKSAENIVIC EKTKVLFGKE PA | 982 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 180 | 606 | 5.6e-136 | 0.9976076555023924 |
| CBM3 | 632 | 715 | 3.2e-16 | 0.9659090909090909 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 1.60e-161 | 183 | 605 | 1 | 374 | Glycosyl hydrolase family 9. |
| PLN00119 | PLN00119 | 7.79e-109 | 174 | 608 | 25 | 488 | endoglucanase |
| PLN02340 | PLN02340 | 5.38e-107 | 179 | 610 | 29 | 495 | endoglucanase |
| PLN02345 | PLN02345 | 1.79e-104 | 184 | 610 | 1 | 460 | endoglucanase |
| PLN02613 | PLN02613 | 1.81e-101 | 169 | 606 | 15 | 476 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABN51281.1 | 1.00e-263 | 177 | 981 | 74 | 887 |
| CAB76932.1 | 1.00e-263 | 177 | 981 | 74 | 887 |
| AAA20892.1 | 6.14e-263 | 177 | 964 | 74 | 863 |
| AEM74912.1 | 7.35e-263 | 179 | 981 | 33 | 839 |
| ADQ41951.1 | 7.35e-263 | 179 | 981 | 33 | 839 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1G87_A | 5.40e-240 | 177 | 781 | 2 | 614 | TheCrystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1G87_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum [Ruminiclostridium cellulolyticum],1GA2_A The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1GA2_B The Crystal Structure Of Endoglucanase 9g From Clostridium Cellulolyticum Complexed With Cellobiose [Ruminiclostridium cellulolyticum],1KFG_A The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum],1KFG_B The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor [Ruminiclostridium cellulolyticum] |
| 1K72_A | 1.74e-238 | 177 | 781 | 2 | 614 | TheX-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum],1K72_B The X-ray Crystal Structure Of Cel9G Complexed With cellotriose [Ruminiclostridium cellulolyticum] |
| 4DOD_A | 1.35e-206 | 177 | 624 | 24 | 475 | Thestructure of Cbescii CelA GH9 module [Caldicellulosiruptor bescii],4DOE_A The liganded structure of Cbescii CelA GH9 module [Caldicellulosiruptor bescii] |
| 2XFG_A | 3.18e-202 | 177 | 612 | 22 | 463 | ChainA, ENDOGLUCANASE 1 [Acetivibrio thermocellus] |
| 1JS4_A | 6.40e-190 | 178 | 781 | 3 | 605 | EndoEXOCELLULASE:CELLOBIOSEFROM THERMOMONOSPORA [Thermobifida fusca],1JS4_B EndoEXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA [Thermobifida fusca],1TF4_A EndoEXOCELLULASE FROM THERMOMONOSPORA [Thermobifida fusca],1TF4_B EndoEXOCELLULASE FROM THERMOMONOSPORA [Thermobifida fusca],3TF4_A EndoEXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA [Thermobifida fusca],3TF4_B EndoEXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA [Thermobifida fusca],4TF4_A EndoEXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA [Thermobifida fusca],4TF4_B EndoEXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02934 | 2.00e-264 | 177 | 981 | 74 | 887 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
| P22534 | 2.71e-251 | 177 | 960 | 24 | 827 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P23659 | 1.96e-242 | 179 | 792 | 30 | 652 | Endoglucanase Z OS=Thermoclostridium stercorarium OX=1510 GN=celZ PE=1 SV=1 |
| P37700 | 1.36e-240 | 177 | 804 | 37 | 672 | Endoglucanase G OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCG PE=1 SV=2 |
| P26224 | 1.04e-232 | 179 | 800 | 30 | 653 | Endoglucanase F OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
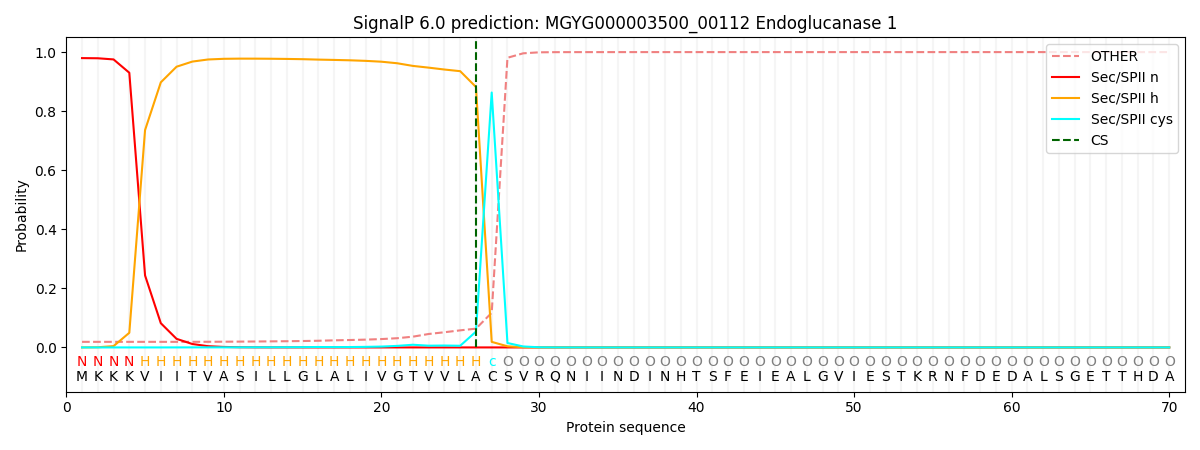
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.018953 | 0.001346 | 0.979699 | 0.000006 | 0.000007 | 0.000010 |