You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003500_00389
You are here: Home > Sequence: MGYG000003500_00389
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Butyrivibrio_A sp900768755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Butyrivibrio_A; Butyrivibrio_A sp900768755 | |||||||||||
| CAZyme ID | MGYG000003500_00389 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47599; End: 48819 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 102 | 371 | 5.5e-98 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.71e-71 | 101 | 372 | 15 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 8.19e-40 | 34 | 398 | 19 | 389 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADK66823.1 | 1.77e-118 | 78 | 400 | 43 | 363 |
| CBL17440.1 | 2.12e-117 | 3 | 400 | 7 | 373 |
| AAF00074.2 | 3.93e-116 | 66 | 400 | 4 | 343 |
| AEX92707.1 | 2.79e-115 | 78 | 400 | 4 | 331 |
| ADQ12983.1 | 2.62e-114 | 81 | 400 | 1 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AYR_A | 2.28e-117 | 66 | 400 | 18 | 357 | GH5endoglucanase EglA from a ruminal fungus [Piromyces rhizinflatus] |
| 3AYS_A | 1.84e-116 | 66 | 400 | 18 | 357 | GH5endoglucanase from a ruminal fungus in complex with cellotriose [Piromyces rhizinflatus] |
| 6Q1I_A | 4.60e-96 | 67 | 399 | 13 | 349 | GH5-4broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum],6Q1I_B GH5-4 broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum] |
| 6WQP_A | 8.38e-96 | 63 | 400 | 11 | 353 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 4W85_A | 1.18e-94 | 70 | 398 | 7 | 334 | Crystalstructure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose [uncultured bacterium],4W85_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from ruminal metagenomic library, in complex with glucose [uncultured bacterium],4W87_A Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide [uncultured bacterium],4W87_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide [uncultured bacterium],4W89_A Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with cellotriose [uncultured bacterium],4W89_B Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with cellotriose [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q12647 | 1.16e-108 | 60 | 399 | 17 | 359 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P23660 | 1.69e-100 | 62 | 399 | 21 | 359 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P23661 | 1.25e-97 | 3 | 400 | 4 | 397 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
| P16216 | 3.22e-97 | 48 | 400 | 48 | 395 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
| P54937 | 3.66e-94 | 67 | 399 | 38 | 374 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
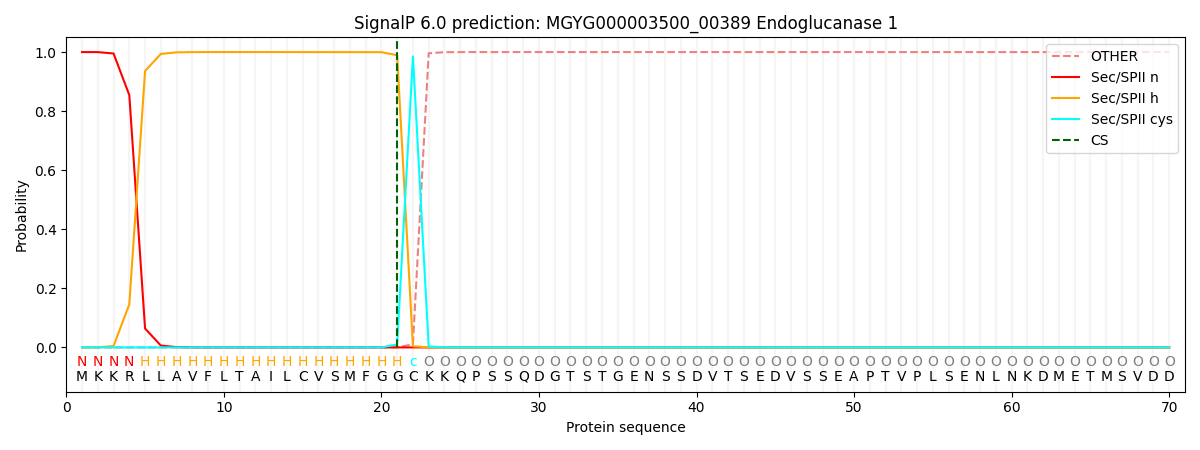
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000047 | 0.000000 | 0.000000 | 0.000000 |
