You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003512_02481
You are here: Home > Sequence: MGYG000003512_02481
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11524 sp900769075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; UBA11524; UBA11524 sp900769075 | |||||||||||
| CAZyme ID | MGYG000003512_02481 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57179; End: 58393 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 33 | 398 | 2.4e-72 | 0.9834983498349835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.24e-66 | 74 | 397 | 1 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.80e-66 | 32 | 399 | 1 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.04e-44 | 2 | 397 | 3 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUZ20632.1 | 8.09e-161 | 1 | 404 | 33 | 436 |
| AHF25185.1 | 5.38e-159 | 3 | 403 | 21 | 425 |
| QUA53142.1 | 8.52e-159 | 1 | 404 | 1 | 408 |
| AHF23772.1 | 6.10e-154 | 1 | 404 | 1 | 408 |
| QTE72954.1 | 2.23e-148 | 1 | 404 | 1 | 408 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3RDK_A | 9.63e-40 | 31 | 397 | 6 | 335 | Proteincrystal structure of xylanase A1 of Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RDK_B Protein crystal structure of xylanase A1 of Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_A Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_B Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_C Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_D Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_E Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_F Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_G Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_H Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],4E4P_A Second native structure of Xylanase A1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],4E4P_B Second native structure of Xylanase A1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2] |
| 7NL2_A | 3.31e-39 | 41 | 374 | 20 | 309 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 6FHE_A | 1.33e-37 | 41 | 397 | 21 | 338 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 2W5F_A | 4.87e-37 | 32 | 401 | 184 | 528 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 3W24_A | 3.37e-36 | 41 | 400 | 17 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23557 | 7.16e-47 | 74 | 377 | 1 | 288 | Putative endo-1,4-beta-xylanase OS=Caldicellulosiruptor saccharolyticus OX=44001 PE=3 SV=1 |
| P26223 | 3.55e-44 | 41 | 397 | 11 | 334 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| Q60037 | 4.61e-41 | 32 | 397 | 370 | 689 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| Q60042 | 1.43e-38 | 32 | 397 | 366 | 685 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q12603 | 1.32e-37 | 11 | 394 | 14 | 346 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
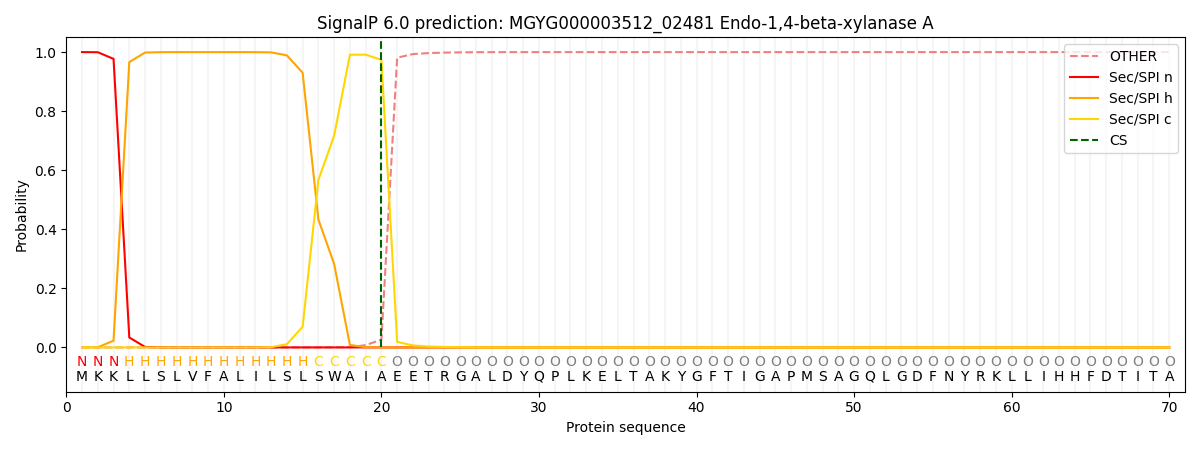
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000303 | 0.998950 | 0.000218 | 0.000171 | 0.000173 | 0.000157 |
