You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003513_01542
You are here: Home > Sequence: MGYG000003513_01542
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900769055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900769055 | |||||||||||
| CAZyme ID | MGYG000003513_01542 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6114; End: 7544 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 206 | 459 | 7.7e-37 | 0.7446153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 4.56e-12 | 218 | 456 | 230 | 452 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 4.20e-11 | 209 | 463 | 88 | 271 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN02218 | PLN02218 | 3.82e-05 | 237 | 441 | 200 | 396 | polygalacturonase ADPG |
| PLN03010 | PLN03010 | 1.34e-04 | 189 | 386 | 115 | 305 | polygalacturonase |
| pfam13229 | Beta_helix | 0.002 | 231 | 391 | 2 | 142 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR43693.1 | 6.23e-155 | 18 | 471 | 27 | 461 |
| QQR19335.1 | 5.77e-153 | 18 | 471 | 27 | 461 |
| ANU59785.1 | 5.77e-153 | 18 | 471 | 27 | 461 |
| QUU00181.1 | 2.18e-151 | 18 | 474 | 21 | 458 |
| ALJ62050.1 | 1.59e-150 | 18 | 471 | 18 | 452 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1HG8_A | 1.86e-07 | 216 | 462 | 114 | 298 | Endopolygalacturonasefrom the phytopathogenic fungus Fusarium moniliforme [Fusarium verticillioides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QTU0 | 3.50e-07 | 209 | 462 | 256 | 447 | Probable endopolygalacturonase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pgaD PE=3 SV=1 |
| Q9P4W2 | 3.50e-07 | 209 | 462 | 256 | 447 | Endopolygalacturonase D OS=Aspergillus niger OX=5061 GN=pgaD PE=3 SV=1 |
| Q07181 | 1.10e-06 | 216 | 462 | 138 | 322 | Polygalacturonase OS=Gibberella fujikuroi OX=5127 GN=PGA PE=1 SV=1 |
| Q873X6 | 1.72e-06 | 215 | 392 | 147 | 346 | Probable exopolygalacturonase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pgxB PE=2 SV=1 |
| A1DAX2 | 1.76e-06 | 215 | 398 | 142 | 347 | Probable exopolygalacturonase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pgxB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
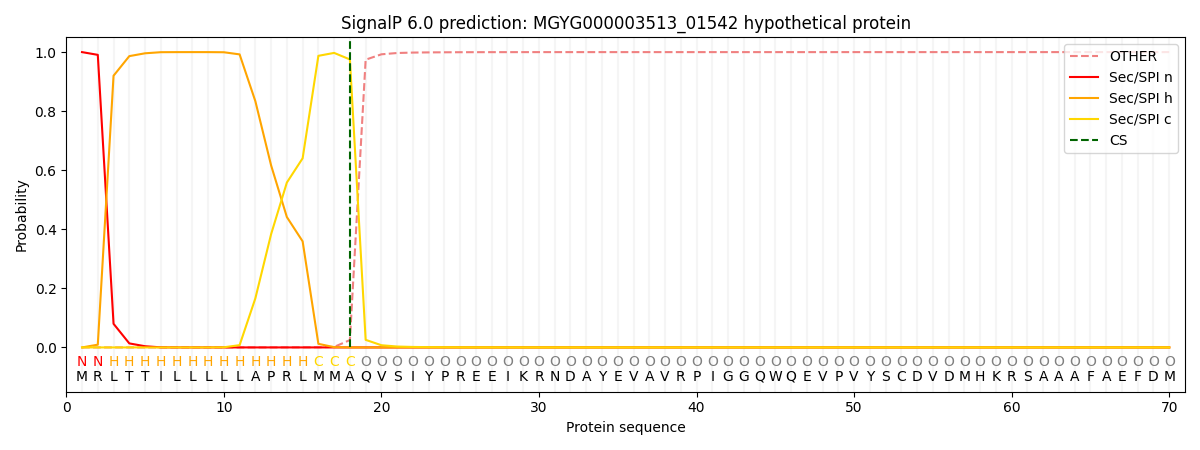
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000318 | 0.998980 | 0.000174 | 0.000175 | 0.000160 | 0.000150 |
