You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003514_02133
You are here: Home > Sequence: MGYG000003514_02133
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fibrobacter_A intestinalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fibrobacterota; Fibrobacteria; Fibrobacterales; Fibrobacteraceae; Fibrobacter_A; Fibrobacter_A intestinalis | |||||||||||
| CAZyme ID | MGYG000003514_02133 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12664; End: 14541 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 36 | 346 | 9.7e-96 | 0.9868852459016394 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13861 | FLgD_tudor | 0.002 | 558 | 621 | 28 | 99 | FlgD Tudor-like domain. This domain has a tudor domain-like beta barrel fold. It is found in the FlgD protein the flagellar hook capping protein. The structure for this domain shows that it contains a nested Ig-like domain within it. However in some firmicute proteins this inserted domain is absent such as Q67K21. |
| pfam13860 | FlgD_ig | 0.003 | 560 | 615 | 13 | 76 | FlgD Ig-like domain. This domains has an immunoglobulin like beta sandwich fold. It is found in the FlgD protein the flagellar hook capping protein. THe structure for this domain shows that it is inserted within a TUDOR like beta barrel domain. |
| TIGR04183 | Por_Secre_tail | 0.004 | 558 | 611 | 8 | 57 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACX74338.1 | 2.03e-304 | 1 | 615 | 1 | 617 |
| AGS52743.1 | 3.00e-159 | 24 | 516 | 70 | 561 |
| AGS52098.1 | 2.72e-69 | 1 | 365 | 1 | 360 |
| AGS53888.1 | 1.49e-68 | 27 | 365 | 30 | 360 |
| AGS52742.1 | 1.28e-61 | 13 | 375 | 11 | 373 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OOU_A | 7.79e-43 | 24 | 345 | 23 | 343 | Crystalstructure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOU_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOZ_A Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus],4OOZ_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus] |
| 2C0H_A | 8.94e-35 | 20 | 356 | 1 | 336 | ChainA, MANNAN ENDO-1,4-BETA-MANNOSIDASE [Mytilus edulis] |
| 3VUP_A | 2.19e-34 | 24 | 358 | 2 | 336 | Beta-1,4-mannanasefrom the common sea hare Aplysia kurodai [Aplysia kurodai],3VUP_B Beta-1,4-mannanase from the common sea hare Aplysia kurodai [Aplysia kurodai] |
| 5Y6T_A | 3.22e-34 | 24 | 347 | 8 | 330 | ChainA, endo-1,4-beta-mannanase [Eisenia fetida] |
| 1QNO_A | 8.81e-08 | 20 | 269 | 1 | 244 | ChainA, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4XC07 | 3.74e-42 | 24 | 345 | 23 | 343 | Mannan endo-1,4-beta-mannosidase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
| Q8WPJ2 | 6.48e-34 | 9 | 356 | 5 | 350 | Mannan endo-1,4-beta-mannosidase OS=Mytilus edulis OX=6550 PE=1 SV=1 |
| Q99036 | 1.28e-08 | 1 | 269 | 1 | 271 | Mannan endo-1,4-beta-mannosidase A OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=man1 PE=1 SV=1 |
| Q5AVP1 | 2.48e-08 | 11 | 382 | 14 | 368 | Mannan endo-1,4-beta-mannosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
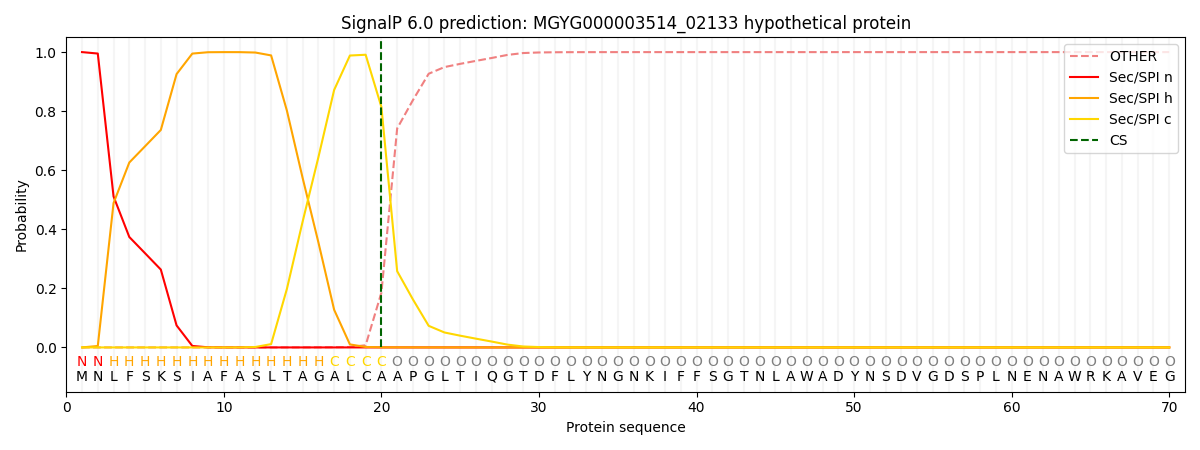
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000355 | 0.998955 | 0.000166 | 0.000184 | 0.000159 | 0.000141 |
