You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003515_00166
You are here: Home > Sequence: MGYG000003515_00166
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900769275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900769275 | |||||||||||
| CAZyme ID | MGYG000003515_00166 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13319; End: 15934 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 63 | 307 | 1.1e-69 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN03080 | PLN03080 | 6.83e-117 | 29 | 829 | 41 | 743 | Probable beta-xylosidase; Provisional |
| PRK15098 | PRK15098 | 2.57e-111 | 86 | 837 | 115 | 734 | beta-glucosidase BglX. |
| COG1472 | BglX | 1.94e-68 | 47 | 409 | 38 | 368 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 1.23e-46 | 380 | 750 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00933 | Glyco_hydro_3 | 3.07e-39 | 77 | 339 | 73 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85397.1 | 0.0 | 23 | 868 | 2 | 838 |
| QNT67604.1 | 0.0 | 22 | 870 | 21 | 866 |
| ADD92014.1 | 0.0 | 20 | 866 | 40 | 880 |
| QUT88841.1 | 0.0 | 16 | 871 | 9 | 862 |
| ALJ60147.1 | 0.0 | 16 | 871 | 9 | 862 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VC7_A | 7.59e-88 | 40 | 859 | 25 | 722 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 7.59e-88 | 40 | 859 | 25 | 722 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 5Z87_A | 1.19e-83 | 55 | 870 | 109 | 784 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 6Q7I_A | 5.64e-75 | 22 | 856 | 34 | 741 | GH3exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
| 5A7M_A | 1.27e-72 | 39 | 864 | 51 | 750 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 0.0 | 19 | 870 | 13 | 861 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| Q9SGZ5 | 4.89e-98 | 39 | 823 | 47 | 725 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
| Q94KD8 | 6.97e-98 | 39 | 870 | 53 | 765 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
| Q9LJN4 | 1.33e-90 | 32 | 859 | 43 | 762 | Probable beta-D-xylosidase 5 OS=Arabidopsis thaliana OX=3702 GN=BXL5 PE=2 SV=2 |
| Q9FGY1 | 1.13e-88 | 38 | 856 | 57 | 763 | Beta-D-xylosidase 1 OS=Arabidopsis thaliana OX=3702 GN=BXL1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
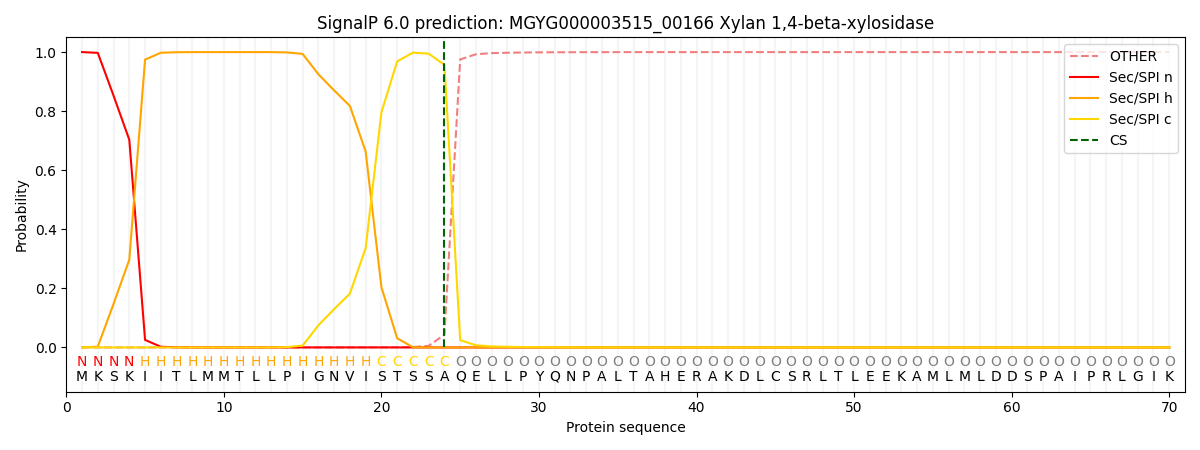
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000283 | 0.999064 | 0.000175 | 0.000167 | 0.000152 | 0.000145 |
