You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003515_00568
You are here: Home > Sequence: MGYG000003515_00568
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900769275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900769275 | |||||||||||
| CAZyme ID | MGYG000003515_00568 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19070; End: 20236 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 32 | 361 | 4.8e-73 | 0.9925650557620818 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVJ82134.1 | 3.71e-157 | 24 | 372 | 44 | 387 |
| ADE83410.1 | 1.73e-155 | 30 | 372 | 51 | 387 |
| QUT89422.1 | 5.39e-130 | 30 | 386 | 55 | 404 |
| ALJ59535.1 | 1.08e-129 | 30 | 386 | 55 | 404 |
| AHW58787.1 | 1.30e-119 | 25 | 388 | 48 | 401 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PIC_A | 6.70e-64 | 45 | 388 | 37 | 369 | ChainA, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
| 6RU2_A | 1.38e-59 | 33 | 386 | 18 | 372 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
| 6RTV_A | 3.62e-59 | 33 | 386 | 24 | 378 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RTV_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RU1_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RU1_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RV7_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV7_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV9_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor],6RV9_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor] |
| 6RV8_A | 8.57e-59 | 33 | 386 | 96 | 450 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
| 4G4G_A | 7.23e-55 | 24 | 385 | 48 | 400 | Crystalstructure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G0RV93 | 3.16e-62 | 45 | 388 | 122 | 454 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
| K5XDZ6 | 5.35e-60 | 33 | 388 | 44 | 399 | 4-O-methyl-glucuronoyl methylesterase OS=Phanerochaete carnosa (strain HHB-10118-sp) OX=650164 GN=PHACADRAFT_247750 PE=1 SV=1 |
| P0CT88 | 5.49e-60 | 33 | 386 | 44 | 392 | 4-O-methyl-glucuronoyl methylesterase 2 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.11.1537.1 PE=1 SV=1 |
| A0A0A7EQR3 | 4.02e-58 | 33 | 386 | 112 | 466 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
| B2ABS0 | 1.81e-57 | 33 | 385 | 123 | 472 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
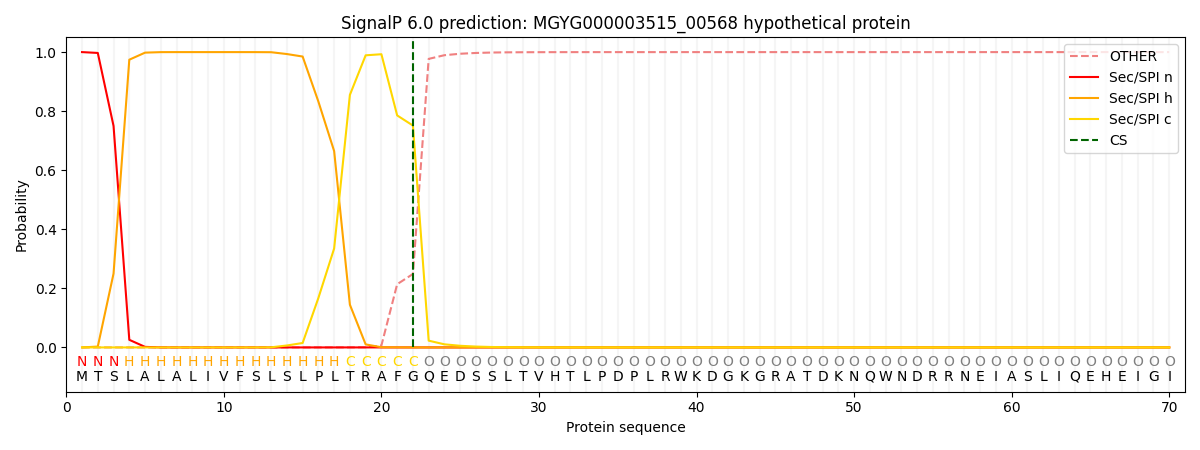
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999117 | 0.000152 | 0.000175 | 0.000156 | 0.000142 |
