You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003520_01552
You are here: Home > Sequence: MGYG000003520_01552
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900769145 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900769145 | |||||||||||
| CAZyme ID | MGYG000003520_01552 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 245; End: 1594 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 60 | 248 | 6e-17 | 0.45864661654135336 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 3.58e-30 | 59 | 413 | 1 | 338 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 3.61e-10 | 62 | 168 | 1 | 96 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| pfam01113 | DapB_N | 2.54e-06 | 62 | 160 | 1 | 90 | Dihydrodipicolinate reductase, N-terminus. Dihydrodipicolinate reductase (DapB) reduces the alpha,beta-unsaturated cyclic imine, dihydro-dipicolinate. This reaction is the second committed step in the biosynthesis of L-lysine and its precursor meso-diaminopimelate, which are critical for both protein and cell wall biosynthesis. The N-terminal domain of DapB binds the dinucleotide NADPH. |
| COG0289 | DapB | 1.33e-04 | 62 | 160 | 3 | 95 | Dihydrodipicolinate reductase [Amino acid transport and metabolism]. |
| PRK00048 | PRK00048 | 2.00e-04 | 62 | 160 | 2 | 86 | dihydrodipicolinate reductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23801.1 | 4.32e-229 | 4 | 449 | 5 | 453 |
| CDN31231.1 | 5.15e-221 | 4 | 449 | 3 | 454 |
| BCG53919.1 | 2.69e-220 | 2 | 449 | 3 | 459 |
| BBK93797.1 | 8.00e-210 | 2 | 447 | 3 | 455 |
| QIX65768.1 | 8.00e-210 | 2 | 447 | 3 | 455 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MKX_A | 5.58e-09 | 56 | 286 | 12 | 218 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4MKZ_A Chain A, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4N54_A | 7.34e-09 | 60 | 286 | 13 | 215 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 1ZH8_A | 2.82e-06 | 60 | 388 | 17 | 309 | Crystalstructure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution [Thermotoga maritima MSB8],1ZH8_B Crystal structure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01S58 | 3.95e-08 | 4 | 248 | 2 | 229 | Glycosyl hydrolase family 109 protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6590 PE=3 SV=1 |
| Q88S38 | 3.93e-07 | 63 | 291 | 8 | 205 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase OS=Lactiplantibacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1) OX=220668 GN=iolG PE=3 SV=1 |
| A4FK61 | 3.69e-06 | 57 | 216 | 1 | 148 | Inositol 2-dehydrogenase 4 OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=iolG4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
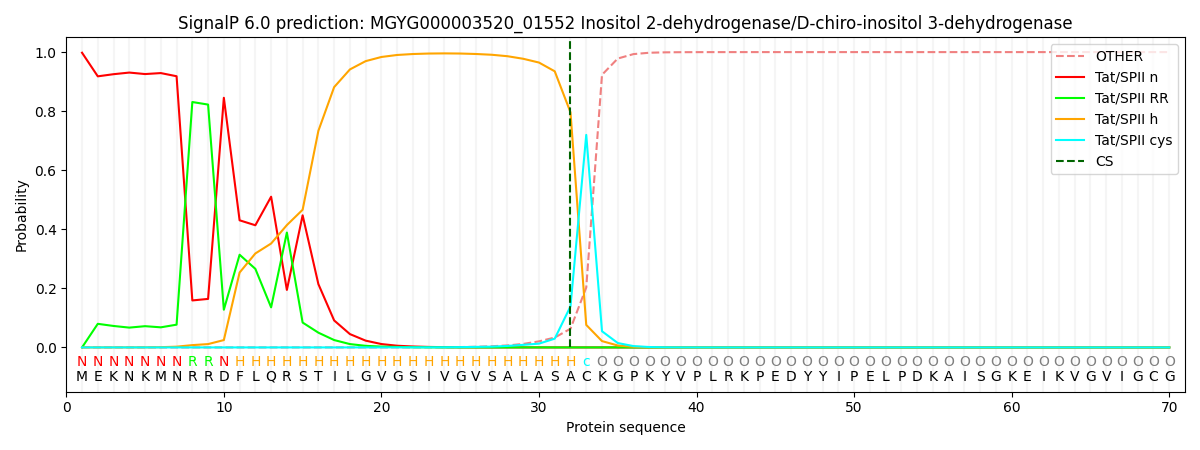
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000019 | 0.002113 | 0.997875 | 0.000000 |
