You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003525_00013
You are here: Home > Sequence: MGYG000003525_00013
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp004556005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp004556005 | |||||||||||
| CAZyme ID | MGYG000003525_00013 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12933; End: 15347 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 501 | 762 | 6.3e-76 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd17332 | MFS_MelB_like | 2.03e-93 | 11 | 433 | 1 | 406 | Salmonella enterica Na+/melibiose symporter MelB and similar transporters of the Major Facilitator Superfamily. This family is composed of Salmonella enterica Na+/melibiose symporter MelB, Major Facilitator Superfamily domain-containing proteins, MFSD2 and MFSD12, and other sugar transporters. MelB catalyzes the electrogenic symport of galactosides with Na+, Li+ or H+. The MFSD2 subfamily is composed of two vertebrate members, MFSD2A and MFSD2B. MFSD2A is more commonly called sodium-dependent lysophosphatidylcholine symporter 1 (NLS1). It is an LPC symporter that plays an essential role for blood-brain barrier formation and function. Inactivating mutations in MFSD2A cause a lethal microcephaly syndrome. MFSD2B is a potential risk or protect factor in the prognosis of lung adenocarcinoma. MelB-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| COG2211 | MelB | 4.43e-89 | 1 | 399 | 4 | 398 | Na+/melibiose symporter or related transporter [Carbohydrate transport and metabolism]. |
| TIGR00792 | gph | 2.73e-72 | 11 | 406 | 1 | 387 | sugar (Glycoside-Pentoside-Hexuronide) transporter. The Glycoside-Pentoside-Hexuronide (GPH):Cation Symporter Family (TC 2.A.2) GPH:cation symporters catalyze uptake of sugars in symport with a monovalent cation (H+ or Na+). Members of this family includes transporters for melibiose, lactose, raffinose, glucuronides, pentosides and isoprimeverose. Mutants of two groups of these symporters (the melibiose permeases of enteric bacteria, and the lactose permease of Streptococcus thermophilus) have been isolated in which altered cation specificity is observed or in which sugar transport is uncoupled from cation symport (i.e., uniport is catalyzed). The various members of the family can use Na+, H+ or Li, Na+ or Li+, H+ or Li+, or only H+ as the symported cation. All of these proteins possess twelve putative transmembrane a-helical spanners. [Transport and binding proteins, Carbohydrates, organic alcohols, and acids] |
| PRK09669 | PRK09669 | 2.64e-67 | 1 | 433 | 1 | 414 | putative symporter YagG; Provisional |
| pfam13347 | MFS_2 | 4.04e-65 | 13 | 399 | 1 | 385 | MFS/sugar transport protein. This family is part of the major facilitator superfamily of membrane transport proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05684.1 | 1.35e-130 | 487 | 803 | 49 | 347 |
| ADU86904.1 | 8.29e-130 | 470 | 803 | 7 | 320 |
| CRY96302.1 | 2.60e-108 | 490 | 804 | 28 | 314 |
| QUT89667.1 | 4.24e-107 | 490 | 804 | 39 | 325 |
| ALJ59275.1 | 1.80e-105 | 490 | 804 | 39 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XZW_A | 3.07e-55 | 490 | 799 | 12 | 299 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 5IHS_A | 4.11e-55 | 490 | 803 | 35 | 321 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 4XZB_A | 8.28e-55 | 490 | 799 | 12 | 300 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 6GJF_A | 1.03e-52 | 490 | 804 | 13 | 302 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 3.53e-51 | 490 | 800 | 37 | 322 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P31435 | 1.30e-52 | 1 | 444 | 1 | 424 | Inner membrane symporter YicJ OS=Escherichia coli (strain K12) OX=83333 GN=yicJ PE=1 SV=2 |
| P75683 | 1.85e-49 | 5 | 438 | 3 | 417 | Putative glycoside/cation symporter YagG OS=Escherichia coli (strain K12) OX=83333 GN=yagG PE=1 SV=1 |
| P10475 | 1.05e-48 | 490 | 800 | 42 | 327 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 2.67e-48 | 490 | 800 | 42 | 327 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P23549 | 1.73e-47 | 490 | 800 | 42 | 327 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000024 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
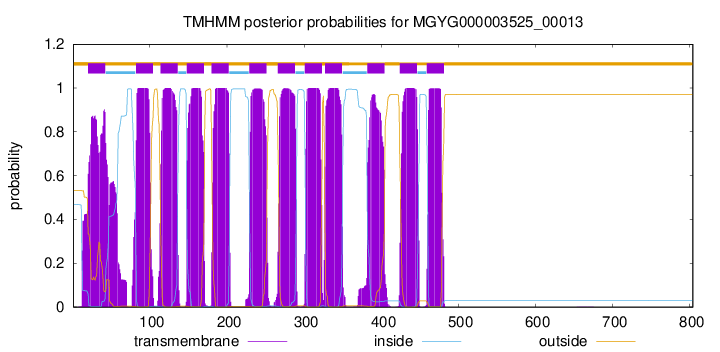
| start | end |
|---|---|
| 20 | 42 |
| 82 | 104 |
| 114 | 136 |
| 148 | 170 |
| 180 | 202 |
| 229 | 251 |
| 266 | 288 |
| 301 | 323 |
| 327 | 349 |
| 382 | 404 |
| 424 | 446 |
| 459 | 481 |
