You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003528_02006
You are here: Home > Sequence: MGYG000003528_02006
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAAGGB01 sp900769285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; UBA1407; UBA1407; CAAGGB01; CAAGGB01 sp900769285 | |||||||||||
| CAZyme ID | MGYG000003528_02006 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11401; End: 14247 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 286 | 659 | 9.2e-59 | 0.5421133231240429 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNN20901.1 | 4.85e-106 | 26 | 929 | 43 | 920 |
| AHF11385.1 | 7.27e-105 | 29 | 656 | 55 | 688 |
| AHF11388.1 | 2.63e-85 | 223 | 656 | 90 | 524 |
| ASI98670.1 | 1.67e-63 | 230 | 660 | 79 | 482 |
| ACS89391.1 | 4.87e-62 | 232 | 659 | 79 | 478 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z6P_A | 6.57e-33 | 270 | 665 | 392 | 762 | Thecrystal structure of an agarase, AgWH50C [Agarivorans gilvus],5Z6P_B The crystal structure of an agarase, AgWH50C [Agarivorans gilvus] |
| 4BQ2_A | 2.97e-26 | 270 | 667 | 373 | 748 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 6.84e-26 | 270 | 667 | 373 | 748 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 6XJ9_A | 1.50e-24 | 252 | 657 | 372 | 754 | Structureof PfGH50B [Pseudoalteromonas fuliginea],6XJ9_B Structure of PfGH50B [Pseudoalteromonas fuliginea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48840 | 2.37e-28 | 184 | 660 | 412 | 947 | Beta-agarase B OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaB PE=3 SV=1 |
| P48839 | 3.06e-22 | 231 | 647 | 455 | 898 | Beta-agarase A OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
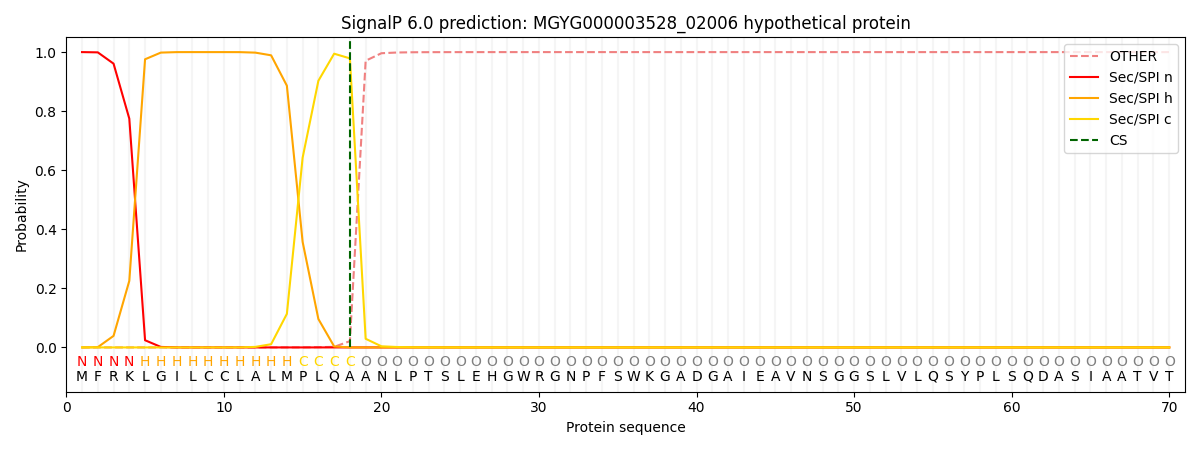
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000266 | 0.998995 | 0.000225 | 0.000169 | 0.000177 | 0.000149 |
