You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003533_02203
You are here: Home > Sequence: MGYG000003533_02203
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
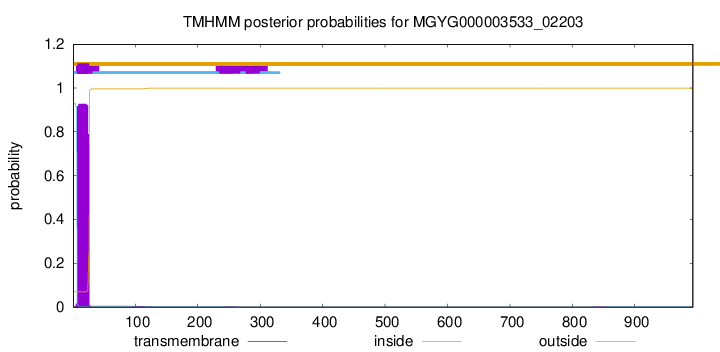
TMHMM annotations
Basic Information help
| Species | SFTJ01 sp900769345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; SFTJ01; SFTJ01 sp900769345 | |||||||||||
| CAZyme ID | MGYG000003533_02203 | |||||||||||
| CAZy Family | CBM57 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1159; End: 4140 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM57 | 533 | 636 | 2.1e-23 | 0.7278911564625851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11721 | Malectin | 6.11e-16 | 510 | 634 | 16 | 149 | Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognizes and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQT67390.1 | 1.58e-227 | 11 | 991 | 12 | 1039 |
| ASV75718.1 | 4.68e-202 | 22 | 987 | 25 | 1008 |
| ACB75512.1 | 6.60e-185 | 4 | 988 | 11 | 1012 |
| QKH85008.1 | 2.73e-20 | 1 | 355 | 1 | 349 |
| QCQ35918.1 | 2.73e-20 | 1 | 355 | 1 | 349 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S7T_I | 4.96e-06 | 508 | 607 | 63 | 165 | Cryo-EMstructure of human oligosaccharyltransferase complex OST-B [Homo sapiens] |
| 2JWP_A | 8.51e-06 | 539 | 607 | 52 | 123 | ChainA, Malectin [unidentified] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
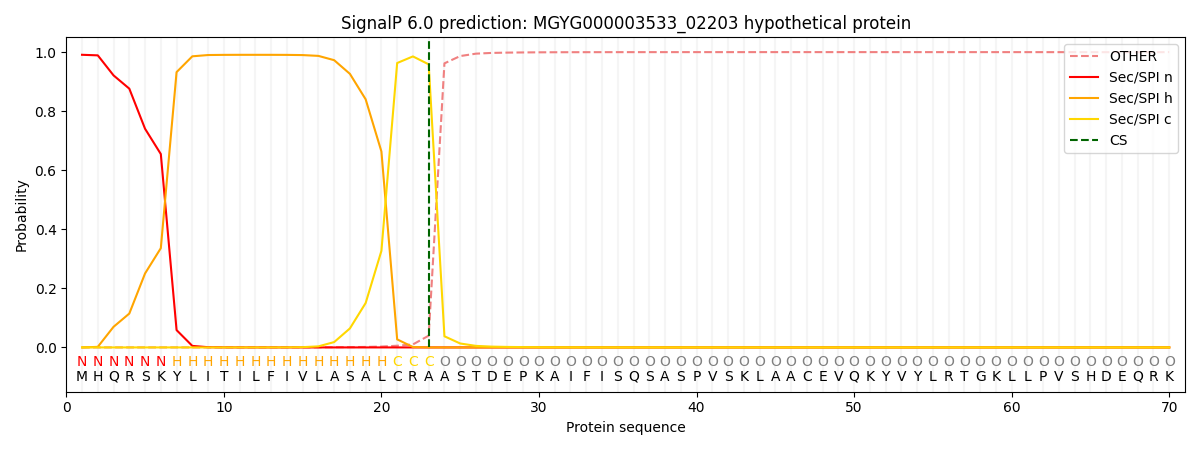
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000817 | 0.987961 | 0.010531 | 0.000254 | 0.000200 | 0.000183 |