You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003535_00532
You are here: Home > Sequence: MGYG000003535_00532
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp004556065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp004556065 | |||||||||||
| CAZyme ID | MGYG000003535_00532 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3165; End: 4385 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 190 | 346 | 7.5e-44 | 0.7178217821782178 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.72e-37 | 197 | 403 | 126 | 340 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 1.35e-36 | 195 | 347 | 39 | 189 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 3.81e-21 | 197 | 344 | 61 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADE83376.1 | 4.96e-108 | 21 | 403 | 42 | 418 |
| QVJ81552.1 | 1.99e-107 | 21 | 403 | 42 | 418 |
| AAW84045.1 | 6.85e-94 | 75 | 403 | 1 | 321 |
| AEW00861.1 | 2.55e-44 | 197 | 398 | 106 | 309 |
| ABG58437.1 | 6.91e-41 | 187 | 402 | 106 | 329 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZSC_A | 5.19e-17 | 171 | 320 | 65 | 213 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 3VMV_A | 2.03e-16 | 197 | 403 | 104 | 321 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1VBL_A | 2.32e-15 | 196 | 403 | 156 | 411 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 1OOC_A | 4.29e-14 | 197 | 390 | 115 | 343 | ChainA, Pectate lyase A [Dickeya chrysanthemi],1OOC_B Chain B, Pectate lyase A [Dickeya chrysanthemi],1PE9_A Chain A, Pectate lyase A [Dickeya chrysanthemi],1PE9_B Chain B, Pectate lyase A [Dickeya chrysanthemi] |
| 1JRG_A | 1.86e-13 | 197 | 390 | 115 | 343 | ChainA, Pectate lyase [Dickeya chrysanthemi],1JRG_B Chain B, Pectate lyase [Dickeya chrysanthemi],1JTA_A Chain A, pectate lyase A [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1L969 | 1.61e-20 | 171 | 320 | 90 | 238 | Pectate trisaccharide-lyase OS=Thermotoga sp. (strain RQ2) OX=126740 GN=pelA PE=3 SV=1 |
| Q9WYR4 | 2.24e-20 | 171 | 320 | 92 | 240 | Pectate trisaccharide-lyase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pelA PE=1 SV=1 |
| Q0CBV0 | 7.24e-17 | 197 | 344 | 115 | 257 | Probable pectate lyase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyB PE=3 SV=1 |
| Q5AVN4 | 1.00e-16 | 196 | 403 | 122 | 322 | Pectate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyA PE=1 SV=1 |
| Q00645 | 1.34e-16 | 197 | 344 | 116 | 258 | Pectate lyase plyB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
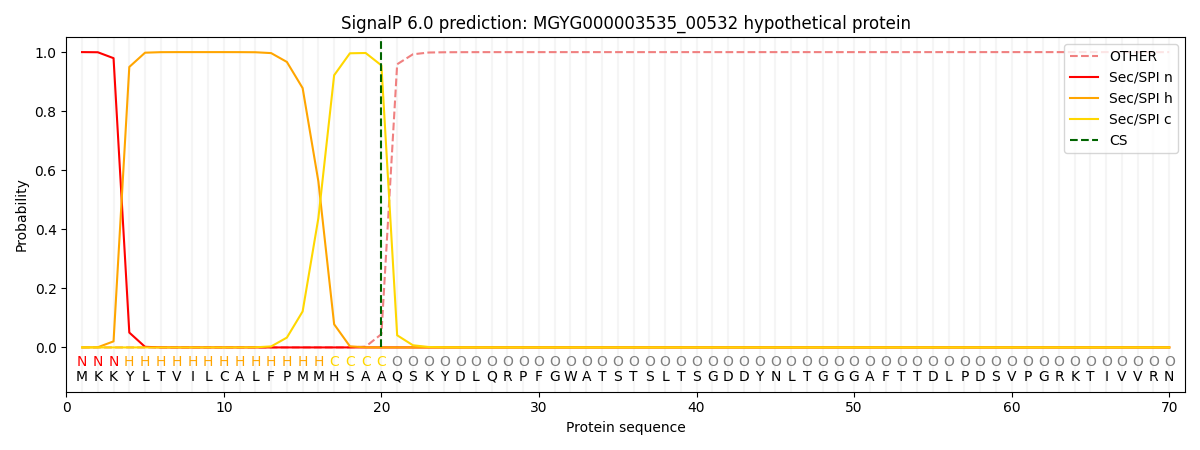
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000359 | 0.998833 | 0.000247 | 0.000194 | 0.000180 | 0.000160 |
