You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003536_00729
You are here: Home > Sequence: MGYG000003536_00729
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; | |||||||||||
| CAZyme ID | MGYG000003536_00729 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1354; End: 3867 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 20 | 835 | 9.9e-211 | 0.9902912621359223 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 3.03e-39 | 128 | 614 | 380 | 853 | alpha-L-rhamnosidase. |
| pfam17132 | Glyco_hydro_106 | 8.72e-05 | 25 | 124 | 6 | 107 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACT94671.1 | 5.13e-212 | 1 | 835 | 1 | 839 |
| QIP13528.1 | 2.99e-211 | 22 | 834 | 30 | 854 |
| QHV95032.1 | 8.47e-208 | 22 | 834 | 30 | 854 |
| AEE52203.1 | 8.54e-208 | 4 | 834 | 21 | 904 |
| AKD54767.1 | 1.28e-205 | 14 | 834 | 27 | 875 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 5.34e-33 | 146 | 837 | 461 | 1141 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 3.78e-21 | 141 | 834 | 412 | 1098 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 8.60e-21 | 141 | 834 | 412 | 1098 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
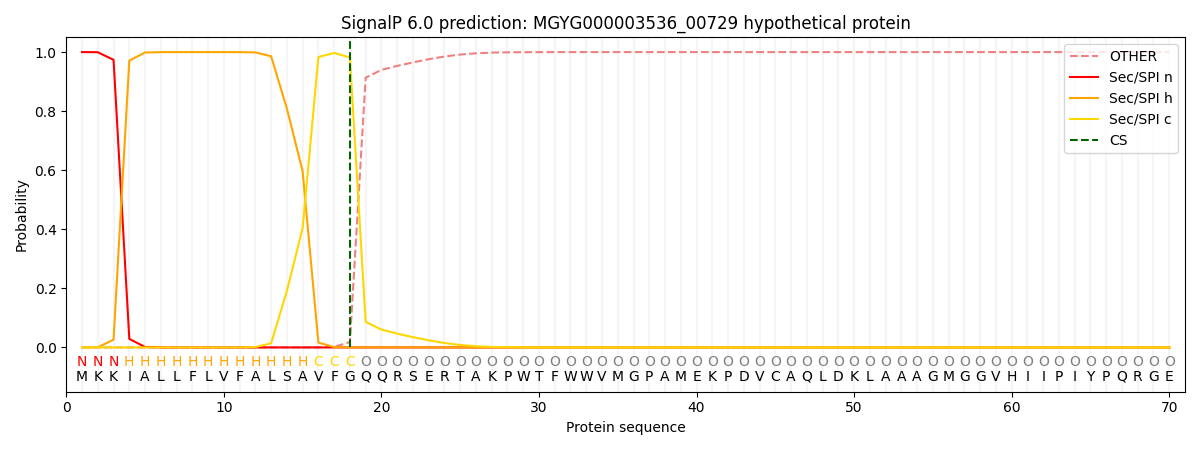
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000299 | 0.999015 | 0.000210 | 0.000164 | 0.000167 | 0.000160 |
