You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003536_01464
You are here: Home > Sequence: MGYG000003536_01464
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; | |||||||||||
| CAZyme ID | MGYG000003536_01464 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1212; End: 2831 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 104 | 489 | 7.2e-42 | 0.9603960396039604 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.79e-35 | 187 | 489 | 17 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 4.13e-30 | 189 | 488 | 62 | 307 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.75e-22 | 171 | 490 | 69 | 338 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA28189.1 | 1.26e-175 | 48 | 529 | 19 | 494 |
| QQZ02681.1 | 1.08e-156 | 53 | 538 | 20 | 494 |
| AVM47074.1 | 1.24e-151 | 47 | 534 | 7 | 491 |
| AHF92621.1 | 1.32e-149 | 57 | 529 | 17 | 481 |
| AWI10666.1 | 2.44e-144 | 58 | 538 | 1 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 1.85e-20 | 188 | 489 | 72 | 338 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 4W8L_A | 2.21e-15 | 188 | 489 | 64 | 341 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 2F8Q_A | 2.41e-12 | 188 | 463 | 67 | 315 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 2FGL_A | 2.42e-12 | 188 | 463 | 68 | 316 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2FGL_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
| 5EB8_A | 2.44e-12 | 188 | 463 | 69 | 317 | Crystalstructure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27],5EB8_B Crystal structure of aromatic mutant (F4W) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O69230 | 5.04e-14 | 188 | 489 | 430 | 707 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| P40944 | 3.65e-13 | 188 | 489 | 414 | 675 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor sp. (strain Rt8B.4) OX=28238 GN=xynA PE=3 SV=1 |
| P40943 | 7.42e-11 | 188 | 463 | 106 | 349 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 PE=1 SV=1 |
| I1S3C6 | 8.75e-11 | 188 | 494 | 140 | 381 | Endo-1,4-beta-xylanase D OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLD PE=1 SV=1 |
| P49942 | 1.14e-10 | 188 | 484 | 90 | 363 | Endo-1,4-beta-xylanase A OS=Bacteroides ovatus OX=28116 GN=xylI PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
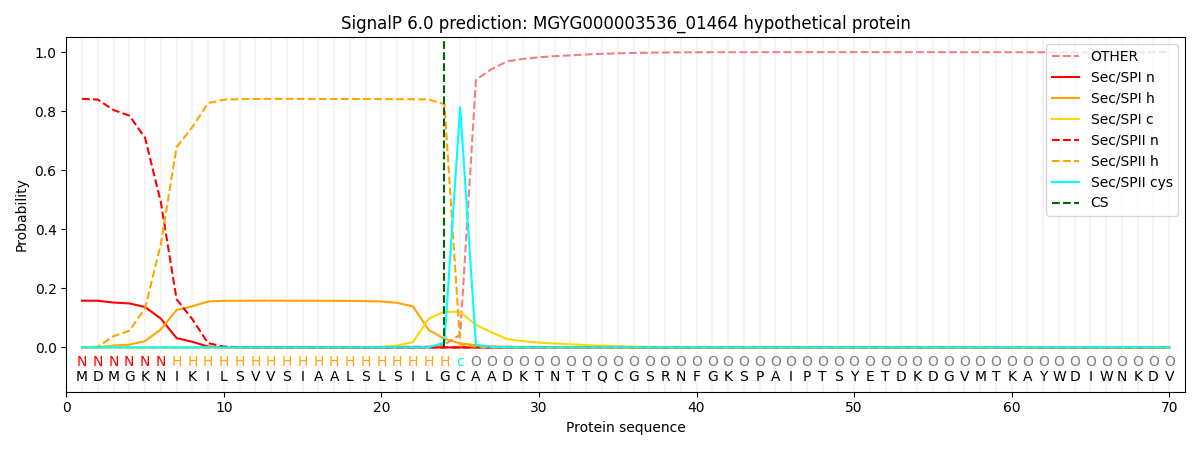
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000290 | 0.154049 | 0.845372 | 0.000104 | 0.000108 | 0.000087 |
