You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003544_00879
You are here: Home > Sequence: MGYG000003544_00879
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900769525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900769525 | |||||||||||
| CAZyme ID | MGYG000003544_00879 | |||||||||||
| CAZy Family | GH53 | |||||||||||
| CAZyme Description | Arabinogalactan endo-beta-1,4-galactanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6357; End: 7325 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 29 | 272 | 3.9e-96 | 0.716374269005848 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07745 | Glyco_hydro_53 | 3.01e-100 | 28 | 318 | 1 | 331 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| COG3867 | GanB | 3.85e-94 | 4 | 320 | 6 | 361 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| cd02932 | OYE_YqiM_FMN | 0.002 | 176 | 236 | 196 | 258 | Old yellow enzyme (OYE) YqjM-like FMN binding domain. YqjM is involved in the oxidative stress response of Bacillus subtilis. Like the other OYE members, each monomer of YqjM contains FMN as a non-covalently bound cofactor and uses NADPH as a reducing agent. The YqjM enzyme exists as a homotetramer that is assembled as a dimer of catalytically dependent dimers, while other OYE members exist only as monomers or dimers. Moreover, the protein displays a shared active site architecture where an arginine finger at the COOH terminus of one monomer extends into the active site of the adjacent monomer and is directly involved in substrate recognition. Another remarkable difference in the binding of the ligand in YqjM is represented by the contribution of the NH2-terminal tyrosine instead of a COOH-terminal tyrosine in OYE and its homologs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AII65942.1 | 1.35e-108 | 24 | 320 | 345 | 639 |
| ARN77257.1 | 1.87e-106 | 18 | 322 | 203 | 505 |
| QKG56411.1 | 5.12e-104 | 11 | 321 | 11 | 328 |
| QDH79868.1 | 7.28e-104 | 25 | 320 | 356 | 650 |
| QJX48698.1 | 3.97e-103 | 13 | 321 | 47 | 359 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7OSK_A | 1.39e-61 | 22 | 270 | 45 | 292 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 6GPA_A | 1.67e-55 | 24 | 320 | 4 | 313 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482],6GPA_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482] |
| 6GP5_A | 4.59e-55 | 24 | 320 | 40 | 349 | Beta-1,4-galactanasefrom Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],6GP5_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 1R8L_A | 7.95e-43 | 21 | 277 | 18 | 270 | Thestructure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1R8L_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1UR0_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR0_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],2CCR_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2CCR_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis] |
| 2GFT_A | 5.92e-42 | 21 | 277 | 18 | 270 | ChainA, Glycosyl Hydrolase Family 53 [Bacillus licheniformis],2GFT_B Chain B, Glycosyl Hydrolase Family 53 [Bacillus licheniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48843 | 2.81e-67 | 24 | 303 | 3 | 312 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| O07013 | 1.90e-44 | 22 | 318 | 48 | 331 | Endo-beta-1,4-galactanase OS=Bacillus subtilis (strain 168) OX=224308 GN=ganB PE=1 SV=1 |
| Q0CTQ7 | 3.64e-44 | 30 | 270 | 20 | 261 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=galA PE=3 SV=1 |
| A1D3T4 | 3.43e-42 | 30 | 270 | 27 | 268 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=galA PE=3 SV=1 |
| Q65CX5 | 7.08e-42 | 21 | 277 | 43 | 295 | Endo-beta-1,4-galactanase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=ganB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
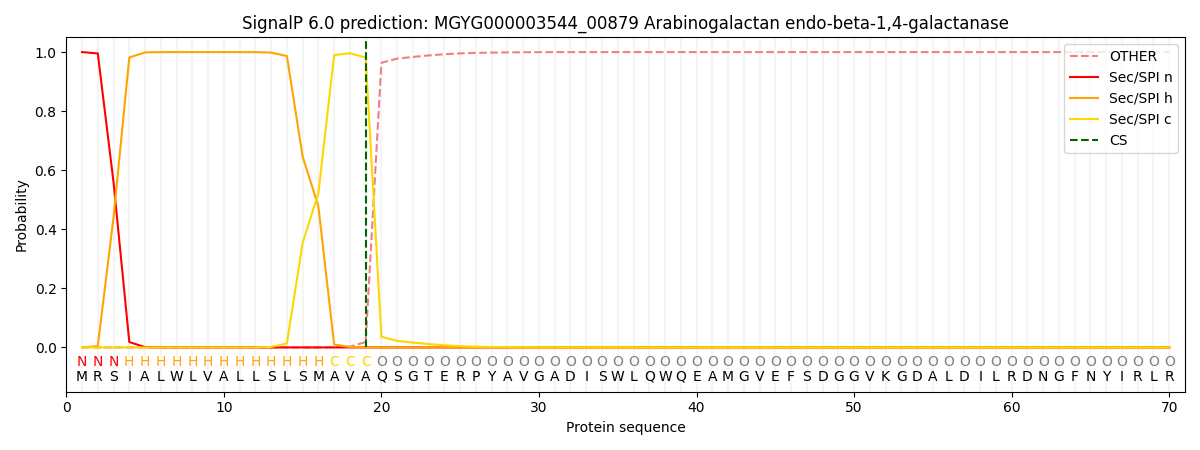
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999085 | 0.000172 | 0.000175 | 0.000161 | 0.000152 |
