You are browsing environment: HUMAN GUT
MGYG000003553_00023
Basic Information
help
Species
HGM05376 sp900769785
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; P3; HGM05376; HGM05376 sp900769785
CAZyme ID
MGYG000003553_00023
CAZy Family
GH163
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
745
85968.08
6.9423
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003553
2321768
MAG
Fiji
Oceania
Gene Location
Start: 1077;
End: 3314
Strand: +
No EC number prediction in MGYG000003553_00023.
Family
Start
End
Evalue
family coverage
GH163
198
446
3.8e-95
0.9840637450199203
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16126
DUF4838
5.38e-107
191
450
1
263
Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown.
more
pfam00754
F5_F8_type_C
4.19e-06
599
727
2
121
F5/8 type C domain. This domain is also known as the discoidin (DS) domain family.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6Q63_A
1.63e-12
582
745
610
774
BT0459[Bacteroides thetaiotaomicron],6Q63_B BT0459 [Bacteroides thetaiotaomicron],6Q63_C BT0459 [Bacteroides thetaiotaomicron]
more
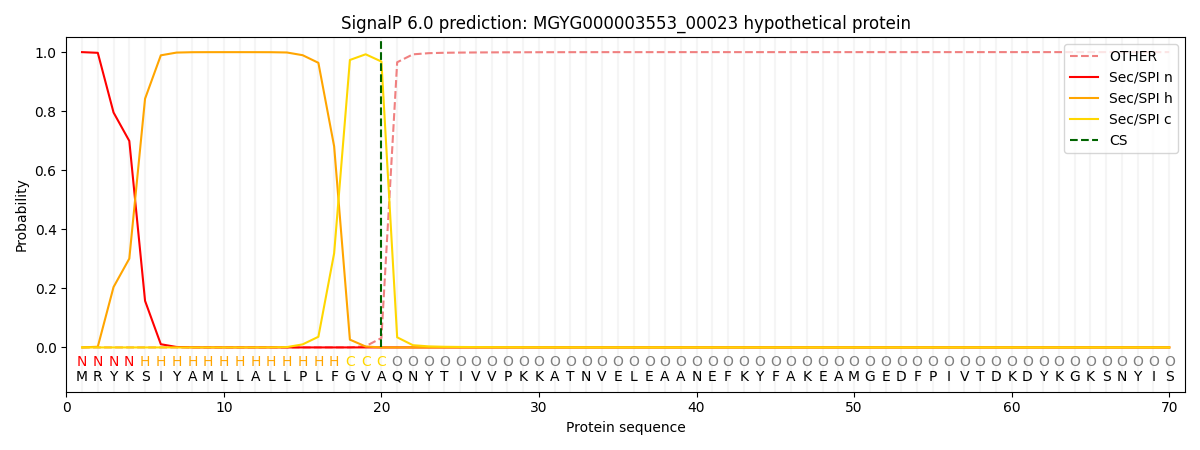
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000458
0.998647
0.000349
0.000167
0.000161
0.000154