You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003556_01264
You are here: Home > Sequence: MGYG000003556_01264
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900769745 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900769745 | |||||||||||
| CAZyme ID | MGYG000003556_01264 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2701; End: 3918 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 33 | 201 | 3e-24 | 0.40100250626566414 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 5.82e-33 | 32 | 315 | 2 | 260 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 3.87e-14 | 34 | 159 | 1 | 110 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 3.34e-04 | 104 | 143 | 57 | 96 | putative oxidoreductase; Provisional |
| COG4091 | COG4091 | 4.49e-04 | 26 | 138 | 10 | 129 | Predicted homoserine dehydrogenase, contains C-terminal SAF domain [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBD45948.1 | 3.99e-129 | 3 | 404 | 6 | 423 |
| BBD45631.1 | 3.41e-124 | 1 | 404 | 1 | 420 |
| CEA16880.1 | 2.04e-120 | 1 | 404 | 1 | 420 |
| SCD19022.1 | 1.16e-119 | 1 | 404 | 1 | 420 |
| QEC68724.1 | 5.37e-116 | 2 | 404 | 5 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EZY_A | 1.94e-15 | 34 | 248 | 3 | 185 | Crystalstructure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima],3EZY_B Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima],3EZY_C Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima],3EZY_D Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima] |
| 3NT2_A | 1.79e-11 | 34 | 192 | 3 | 147 | Crystalstructure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor [Bacillus subtilis],3NT2_B Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor [Bacillus subtilis],3NT4_A Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol [Bacillus subtilis],3NT4_B Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol [Bacillus subtilis],3NT5_A Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose [Bacillus subtilis],3NT5_B Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose [Bacillus subtilis] |
| 2GLX_A | 2.98e-11 | 86 | 264 | 37 | 204 | ChainA, 1,5-anhydro-D-fructose reductase [Ensifer adhaerens],2GLX_B Chain B, 1,5-anhydro-D-fructose reductase [Ensifer adhaerens],2GLX_C Chain C, 1,5-anhydro-D-fructose reductase [Ensifer adhaerens],2GLX_D Chain D, 1,5-anhydro-D-fructose reductase [Ensifer adhaerens],2GLX_E Chain E, 1,5-anhydro-D-fructose reductase [Ensifer adhaerens],2GLX_F Chain F, 1,5-anhydro-D-fructose reductase [Ensifer adhaerens] |
| 4L9R_A | 5.50e-11 | 34 | 192 | 3 | 147 | CrystalStructure of apo A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 4L8V_A | 5.50e-11 | 34 | 192 | 3 | 147 | CrystalStructure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168],4L8V_B Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168],4L8V_C Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168],4L8V_D Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9WYP5 | 1.31e-14 | 34 | 248 | 1 | 183 | Myo-inositol 2-dehydrogenase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=iolG PE=1 SV=1 |
| O05389 | 5.36e-11 | 30 | 273 | 1 | 214 | Uncharacterized oxidoreductase YrbE OS=Bacillus subtilis (strain 168) OX=224308 GN=yrbE PE=3 SV=2 |
| P26935 | 9.78e-11 | 34 | 192 | 3 | 147 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase OS=Bacillus subtilis (strain 168) OX=224308 GN=iolG PE=1 SV=2 |
| Q01S58 | 1.42e-10 | 2 | 245 | 5 | 249 | Glycosyl hydrolase family 109 protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6590 PE=3 SV=1 |
| Q2I8V6 | 1.64e-10 | 86 | 264 | 38 | 205 | 1,5-anhydro-D-fructose reductase OS=Ensifer adhaerens OX=106592 GN=afr PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
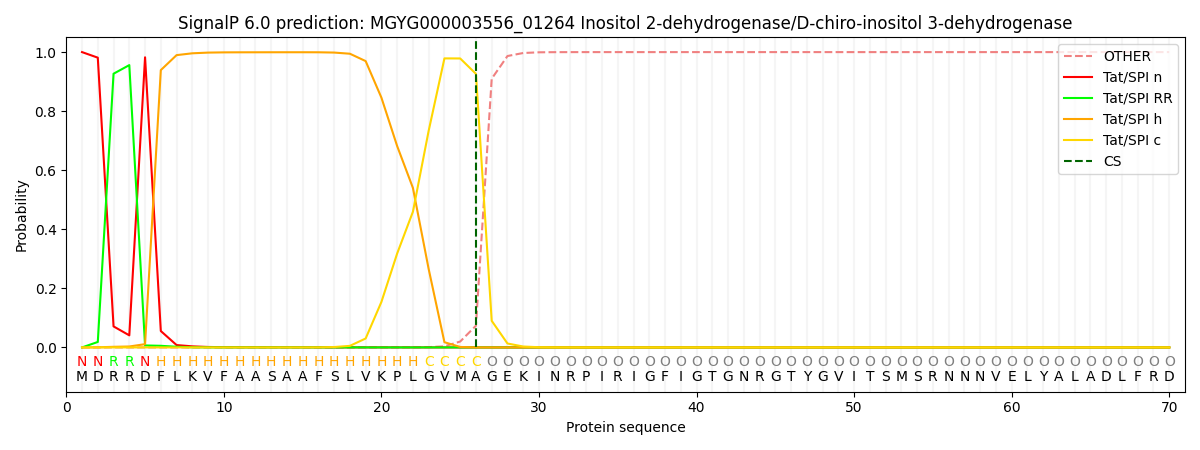
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 1.000011 | 0.000000 | 0.000000 |
