You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003569_00999
You are here: Home > Sequence: MGYG000003569_00999
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
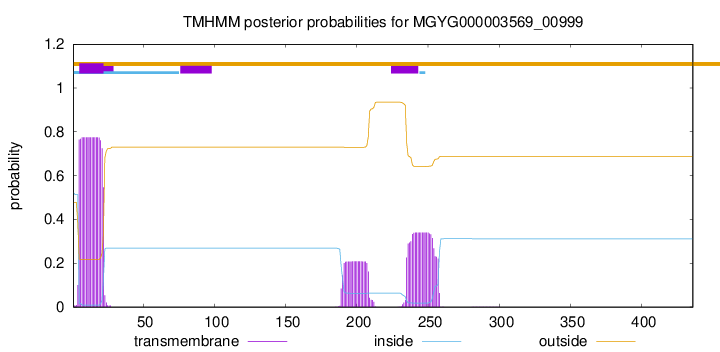
TMHMM annotations
Basic Information help
| Species | Treponema_D sp900769975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Spirochaetota; Spirochaetia; Treponematales; Treponemataceae; Treponema_D; Treponema_D sp900769975 | |||||||||||
| CAZyme ID | MGYG000003569_00999 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8422; End: 9732 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 157 | 298 | 9.1e-22 | 0.9259259259259259 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16894 | MltD-like | 2.07e-52 | 162 | 290 | 1 | 129 | Membrane-bound lytic murein transglycosylase D and similar proteins. Lytic transglycosylases (LT) catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc). Membrane-bound lytic murein transglycosylase D protein (MltD) family members may have one or more small LysM domains, which may contribute to peptidoglycan binding. Unlike the similar "goose-type" lysozymes, LTs also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| PRK10783 | mltD | 2.32e-42 | 116 | 435 | 64 | 389 | membrane-bound lytic murein transglycosylase D; Provisional |
| pfam01464 | SLT | 6.22e-21 | 168 | 268 | 12 | 114 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd00254 | LT-like | 3.70e-14 | 177 | 287 | 10 | 108 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd00118 | LysM | 1.76e-12 | 391 | 433 | 3 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL96994.1 | 3.14e-114 | 124 | 436 | 143 | 455 |
| QGT51390.1 | 3.20e-108 | 91 | 435 | 54 | 396 |
| AEE16302.1 | 2.78e-95 | 106 | 434 | 101 | 419 |
| QTQ11115.1 | 2.84e-89 | 124 | 382 | 80 | 339 |
| QTQ14726.1 | 2.84e-89 | 124 | 382 | 80 | 339 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AEZ7 | 3.55e-25 | 116 | 435 | 61 | 386 | Membrane-bound lytic murein transglycosylase D OS=Escherichia coli (strain K12) OX=83333 GN=mltD PE=1 SV=1 |
| P0AEZ8 | 3.55e-25 | 116 | 435 | 61 | 386 | Membrane-bound lytic murein transglycosylase D OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=mltD PE=3 SV=1 |
| P32820 | 1.10e-08 | 151 | 260 | 22 | 137 | Putative tributyltin chloride resistance protein OS=Alteromonas sp. (strain M-1) OX=29457 GN=tbtA PE=3 SV=1 |
| O07532 | 1.20e-06 | 325 | 436 | 178 | 286 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| O31852 | 2.46e-06 | 324 | 435 | 160 | 270 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
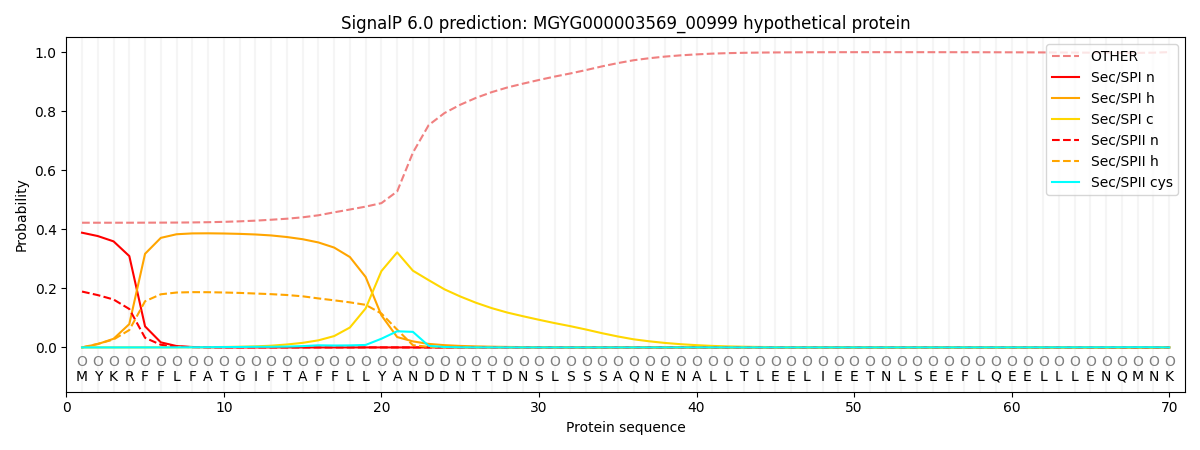
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.436208 | 0.370436 | 0.191881 | 0.000425 | 0.000343 | 0.000694 |