You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003572_00621
You are here: Home > Sequence: MGYG000003572_00621
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900770025 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900770025 | |||||||||||
| CAZyme ID | MGYG000003572_00621 | |||||||||||
| CAZy Family | GH137 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25387; End: 29427 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH137 | 34 | 381 | 2.8e-146 | 0.9941176470588236 |
| GH2 | 389 | 850 | 5.7e-74 | 0.535904255319149 |
| CBM57 | 1040 | 1198 | 1.2e-35 | 0.9863945578231292 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.49e-37 | 397 | 798 | 49 | 426 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam11721 | Malectin | 8.92e-23 | 1038 | 1189 | 1 | 144 | Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognizes and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. |
| PRK10340 | ebgA | 2.65e-20 | 435 | 805 | 112 | 472 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 3.08e-19 | 432 | 799 | 65 | 443 | beta-D-glucuronidase; Provisional |
| pfam02836 | Glyco_hydro_2_C | 1.25e-14 | 663 | 864 | 5 | 226 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCV09658.1 | 0.0 | 34 | 1346 | 20 | 1399 |
| QRQ56950.1 | 0.0 | 34 | 1346 | 20 | 1399 |
| ALJ45323.1 | 0.0 | 34 | 1346 | 20 | 1399 |
| QUT23622.1 | 0.0 | 34 | 1346 | 20 | 1396 |
| QRN01915.1 | 0.0 | 34 | 1346 | 20 | 1399 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MUI_A | 2.96e-179 | 34 | 385 | 26 | 378 | Glycosidehydrolase BT_0996 [Bacteroides thetaiotaomicron VPI-5482] |
| 5MT2_A | 1.31e-172 | 34 | 385 | 26 | 378 | Glycosidehydrolase BT_0996 [Bacteroides thetaiotaomicron VPI-5482],5MUJ_A BT0996 RGII Chain B Complex [Bacteroides thetaiotaomicron VPI-5482] |
| 3CMG_A | 2.79e-47 | 390 | 1019 | 4 | 653 | Crystalstructure of putative beta-galactosidase from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 5Z1A_A | 3.31e-47 | 390 | 1019 | 23 | 672 | Thecrystal structure of Bacteroides fragilis beta-glucuronidase in complex with uronic isofagomine [Bacteroides fragilis NCTC 9343] |
| 6D50_A | 1.83e-42 | 381 | 1185 | 24 | 868 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM09 | 6.94e-33 | 389 | 1012 | 45 | 700 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| A7LXS9 | 8.15e-30 | 396 | 777 | 68 | 437 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| P26257 | 1.20e-22 | 446 | 975 | 69 | 587 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| P77989 | 2.03e-21 | 374 | 971 | 1 | 574 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P06864 | 5.04e-21 | 399 | 778 | 80 | 450 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
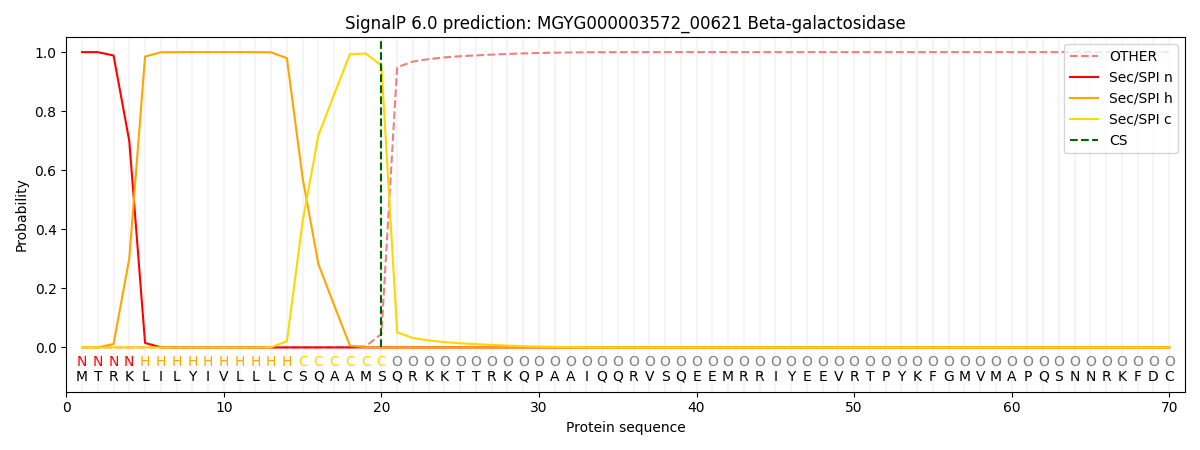
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000257 | 0.999132 | 0.000146 | 0.000154 | 0.000136 | 0.000133 |
