You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003572_01054
You are here: Home > Sequence: MGYG000003572_01054
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
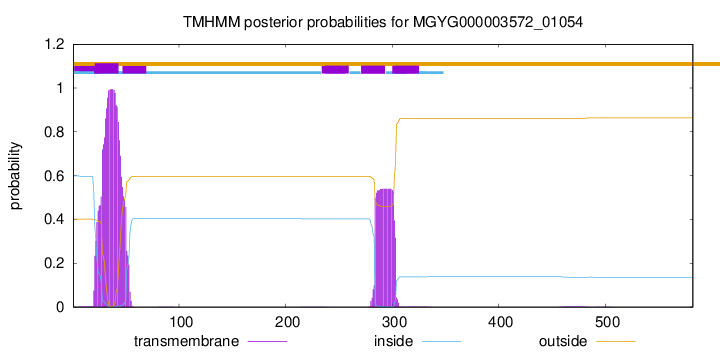
TMHMM annotations
Basic Information help
| Species | Prevotella sp900770025 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900770025 | |||||||||||
| CAZyme ID | MGYG000003572_01054 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54988; End: 56736 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12710 | HAD | 7.88e-33 | 8 | 198 | 2 | 186 | haloacid dehalogenase-like hydrolase. |
| cd03801 | GT4_PimA-like | 8.60e-31 | 213 | 575 | 1 | 366 | phosphatidyl-myo-inositol mannosyltransferase. This family is most closely related to the GT4 family of glycosyltransferases and named after PimA in Propionibacterium freudenreichii, which is involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIM) which are early precursors in the biosynthesis of lipomannans (LM) and lipoarabinomannans (LAM), and catalyzes the addition of a mannosyl residue from GDP-D-mannose (GDP-Man) to the position 2 of the carrier lipid phosphatidyl-myo-inositol (PI) to generate a phosphatidyl-myo-inositol bearing an alpha-1,2-linked mannose residue (PIM1). Glycosyltransferases catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. The acceptor molecule can be a lipid, a protein, a heterocyclic compound, or another carbohydrate residue. This group of glycosyltransferases is most closely related to the previously defined glycosyltransferase family 1 (GT1). The members of this family may transfer UDP, ADP, GDP, or CMP linked sugars. The diverse enzymatic activities among members of this family reflect a wide range of biological functions. The protein structure available for this family has the GTB topology, one of the two protein topologies observed for nucleotide-sugar-dependent glycosyltransferases. GTB proteins have distinct N- and C- terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. The members of this family are found mainly in certain bacteria and archaea. |
| cd02612 | HAD_PGPPase | 1.55e-29 | 7 | 206 | 2 | 191 | phosphatidylglycerol-phosphate phosphatase, similar to Escherichia coli K-12 phosphatidylglycerol-phosphate phosphatase C. This family includes Escherichia coli K-12 phosphatidylglycerol-phosphate phosphatase C, PgpC (previously named yfhB) which catalyzes the dephosphorylation of phosphatidylglycerol-phosphate (PGP) to phosphatidylglycerol (PG). This family belongs to the haloacid dehalogenase-like (HAD) hydrolases, a large superfamily of diverse enzymes that catalyze carbon or phosphoryl group transfer reactions on a range of substrates, using an active site aspartate in nucleophilic catalysis. Members of this superfamily include 2-L-haloalkanoic acid dehalogenase, azetidine hydrolase, phosphonoacetaldehyde hydrolase, phosphoserine phosphatase, phosphomannomutase, P-type ATPases and many others. HAD hydrolases are found in all three kingdoms of life, and most genomes are predicted to contain multiple HAD-like proteins. Members possess a highly conserved alpha/beta core domain, and many also possess a small cap domain, the fold and function of which is variable. HAD hydrolases are sometimes referred to as belonging to the DDDD superfamily of phosphohydrolases. |
| cd03807 | GT4_WbnK-like | 2.38e-26 | 218 | 575 | 4 | 362 | Shigella dysenteriae WbnK and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. WbnK in Shigella dysenteriae has been shown to be involved in the type 7 O-antigen biosynthesis. |
| TIGR01490 | HAD-SF-IB-hyp1 | 3.65e-26 | 6 | 206 | 1 | 195 | HAD-superfamily subfamily IB hydrolase, TIGR01490. This hypothetical equivalog is a member of the IB subfamily (TIGR01488) of the haloacid dehalogenase (HAD) superfamily of aspartate-nucleophile hydrolases. The sequences modelled here are all bacterial. The IB subfamily includes the enzyme phosphoserine phosphatase (TIGR00338). Due to this relationship, several of these sequences have been annotated as "phosphoserine phosphatase related proteins," or "Phosphoserine phosphatase-family enzymes." There is presently no evidence that any of the enzymes in this model possess PSPase activity. OMNI|NTL01ML1250 is annotated as a "possible transferase," however this is due to the C-terminal domain found on this sequence which is homologous to a group of glycerol-phosphate acyltransferases (between trusted and noise to TIGR00530). A subset of these sequences including OMNI|CC1962, the Caulobacter crescentus CicA protein cluster together and may represent a separate equivalog. [Unknown function, Enzymes of unknown specificity] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH14967.1 | 7.70e-135 | 217 | 573 | 7 | 363 |
| QUT46532.1 | 1.28e-108 | 214 | 575 | 1 | 356 |
| QUT35907.1 | 1.73e-105 | 214 | 576 | 1 | 356 |
| QQA28752.1 | 1.73e-105 | 214 | 576 | 1 | 356 |
| QCQ55758.1 | 1.66e-98 | 214 | 575 | 2 | 358 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8GX15 | 6.92e-20 | 6 | 212 | 27 | 220 | Protein CicA OS=Caulobacter vibrioides (strain NA1000 / CB15N) OX=565050 GN=cicA PE=4 SV=1 |
| P0CAV6 | 6.92e-20 | 6 | 212 | 27 | 220 | Protein CicA OS=Caulobacter vibrioides (strain ATCC 19089 / CB15) OX=190650 GN=cicA PE=4 SV=1 |
| Q48453 | 1.90e-06 | 408 | 517 | 191 | 298 | Uncharacterized 41.2 kDa protein in cps region OS=Klebsiella pneumoniae OX=573 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
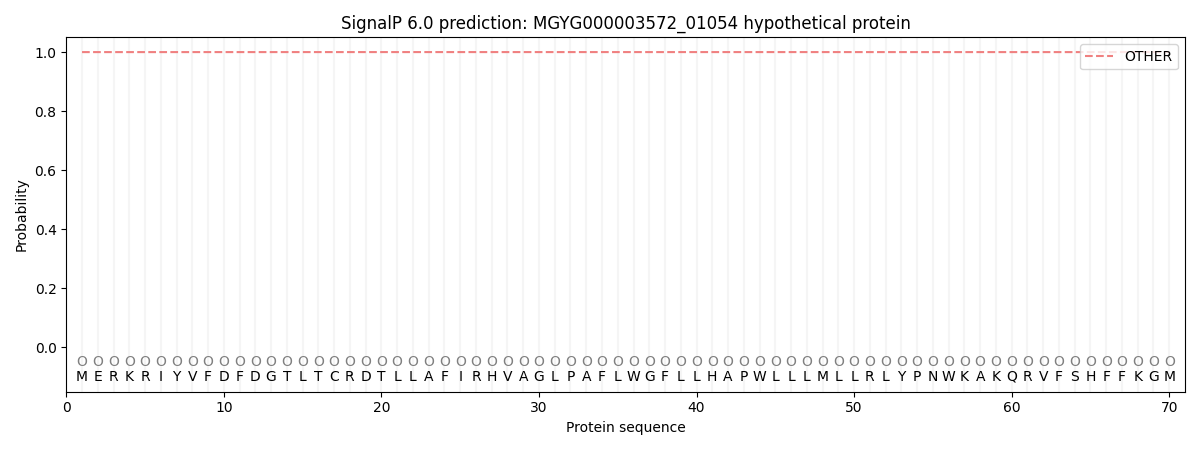
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000024 | 0.000023 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |