You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003581_01018
Basic Information
help
| Species |
HGM12814 sp900770165
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; HGM12814; HGM12814 sp900770165
|
| CAZyme ID |
MGYG000003581_01018
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 336 |
|
38813.06 |
4.8447 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003581 |
2356172 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 336;
End: 1346
Strand: -
|
No EC number prediction in MGYG000003581_01018.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
70 |
309 |
5.7e-28 |
0.9541984732824428 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
5.63e-15 |
73 |
294 |
29 |
257 |
Cellulase (glycosyl hydrolase family 5). |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6KDD_A
|
3.32e-14 |
46 |
331 |
15 |
317 |
endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P25472
|
1.11e-16 |
75 |
271 |
63 |
263 |
Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
This protein is predicted as LIPO
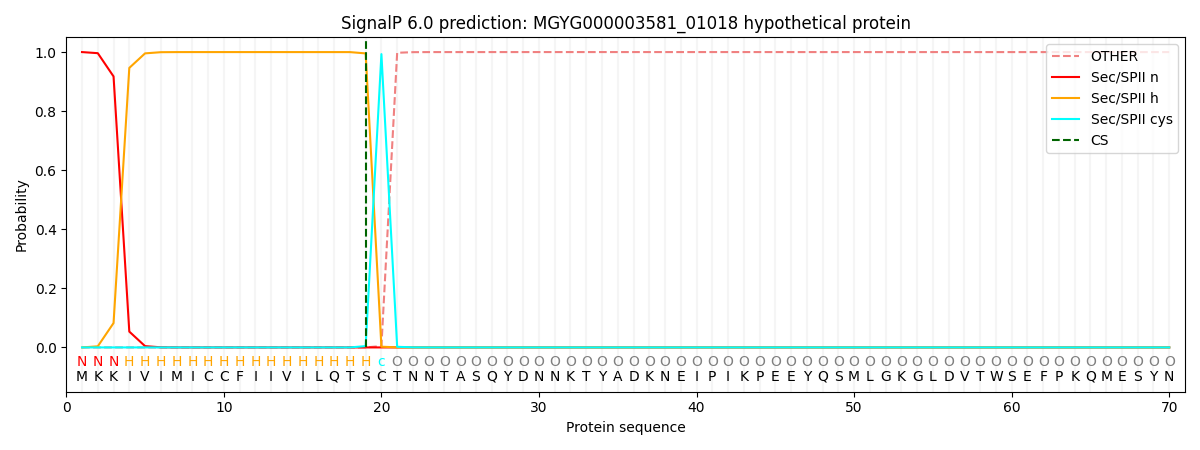
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000000
|
0.000000
|
1.000073
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000003581_01018.

