You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003583_00107
You are here: Home > Sequence: MGYG000003583_00107
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp900770195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp900770195 | |||||||||||
| CAZyme ID | MGYG000003583_00107 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 105252; End: 107150 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 193 | 536 | 9e-96 | 0.9900990099009901 |
| CBM22 | 39 | 164 | 4.2e-23 | 0.9694656488549618 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 2.83e-99 | 193 | 536 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.86e-94 | 234 | 534 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.41e-59 | 213 | 536 | 46 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02018 | CBM_4_9 | 5.08e-16 | 39 | 169 | 3 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| cd14256 | Dockerin_I | 4.59e-13 | 570 | 624 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16579.1 | 5.59e-204 | 1 | 600 | 1 | 598 |
| CAB65753.1 | 4.35e-196 | 37 | 539 | 31 | 533 |
| CAL91979.1 | 1.09e-189 | 40 | 539 | 29 | 526 |
| ADU21885.1 | 9.43e-188 | 28 | 543 | 28 | 545 |
| CAL91978.1 | 4.55e-168 | 69 | 539 | 1 | 466 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5F_A | 2.92e-90 | 60 | 534 | 43 | 524 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 2.54e-87 | 60 | 534 | 43 | 524 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6FHE_A | 6.21e-60 | 189 | 535 | 9 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 7NL2_A | 1.77e-58 | 183 | 546 | 2 | 349 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 5OFJ_A | 3.51e-54 | 181 | 536 | 2 | 337 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29126 | 1.82e-92 | 193 | 531 | 630 | 946 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| P51584 | 6.72e-86 | 60 | 534 | 54 | 535 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q60037 | 1.73e-67 | 39 | 535 | 205 | 690 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| Q60042 | 3.14e-67 | 39 | 535 | 200 | 686 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| P26223 | 1.50e-60 | 193 | 536 | 3 | 336 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
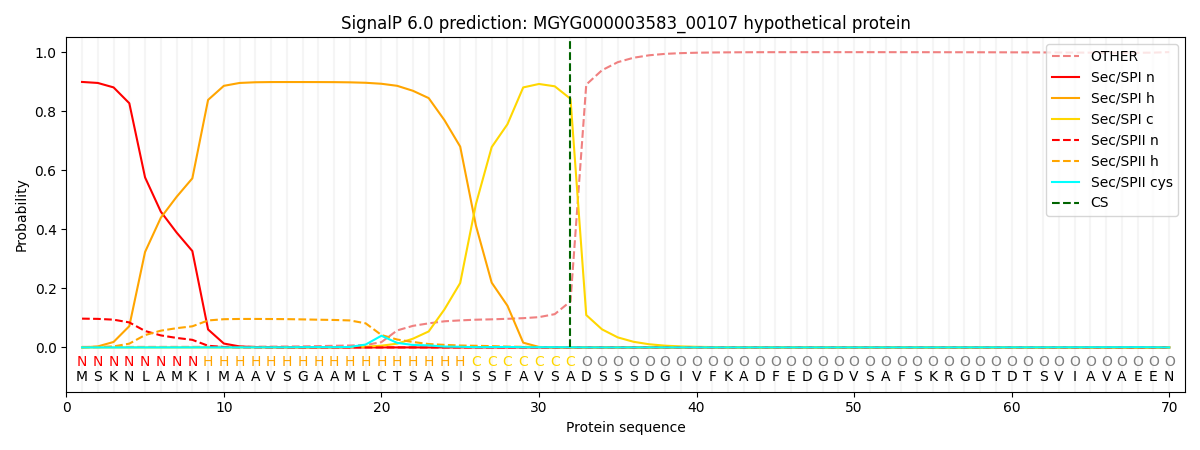
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003746 | 0.890104 | 0.102657 | 0.002679 | 0.000466 | 0.000301 |
