You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003583_01057
You are here: Home > Sequence: MGYG000003583_01057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
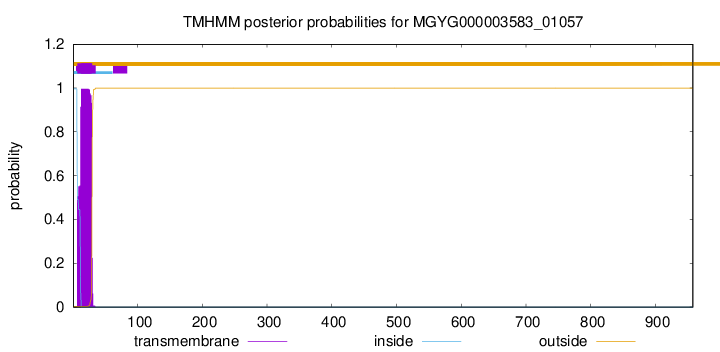
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp900770195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp900770195 | |||||||||||
| CAZyme ID | MGYG000003583_01057 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 93707; End: 96583 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 266 | 450 | 2.7e-55 | 0.9076923076923077 |
| CBM13 | 721 | 878 | 2.2e-21 | 0.7925531914893617 |
| CBM13 | 566 | 715 | 2e-20 | 0.723404255319149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 7.11e-38 | 176 | 522 | 17 | 341 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam14200 | RicinB_lectin_2 | 9.54e-20 | 700 | 794 | 1 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00544 | Pec_lyase_C | 5.31e-19 | 235 | 447 | 1 | 208 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| smart00656 | Amb_all | 2.41e-17 | 262 | 450 | 10 | 186 | Amb_all domain. |
| pfam14200 | RicinB_lectin_2 | 3.16e-17 | 652 | 749 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16867.1 | 0.0 | 37 | 937 | 33 | 892 |
| CDM68184.1 | 5.70e-121 | 40 | 624 | 29 | 584 |
| CDM70399.1 | 2.17e-107 | 40 | 710 | 31 | 697 |
| QHZ48014.1 | 4.24e-98 | 35 | 531 | 26 | 484 |
| VEB20252.1 | 8.25e-98 | 37 | 523 | 28 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZSC_A | 6.76e-11 | 202 | 444 | 14 | 226 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 5B2H_A | 1.79e-09 | 635 | 761 | 163 | 280 | Crystalstructure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum],5B2H_B Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94449 | 3.97e-63 | 187 | 524 | 33 | 338 | Pectin lyase OS=Bacillus subtilis OX=1423 GN=pelB PE=1 SV=1 |
| O34819 | 2.32e-60 | 187 | 524 | 33 | 338 | Pectin lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pelB PE=3 SV=1 |
| P27027 | 8.20e-48 | 189 | 523 | 10 | 304 | Pectin lyase OS=Pseudomonas marginalis OX=298 GN=pnl PE=1 SV=2 |
| P24112 | 6.74e-40 | 186 | 523 | 7 | 306 | Pectin lyase OS=Pectobacterium carotovorum OX=554 GN=pnl PE=1 SV=1 |
| Q00645 | 5.82e-11 | 195 | 427 | 39 | 234 | Pectate lyase plyB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
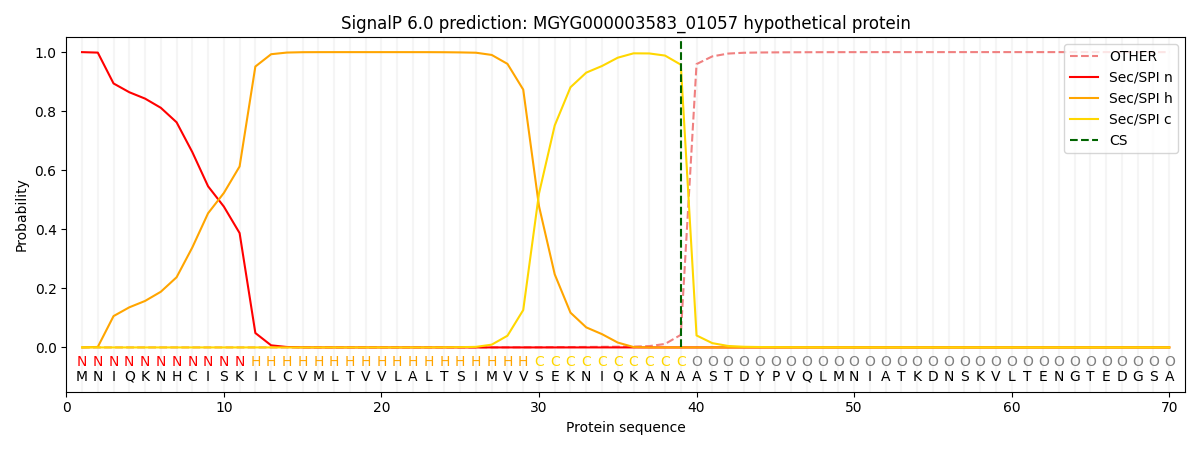
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000352 | 0.998776 | 0.000398 | 0.000156 | 0.000152 | 0.000144 |