You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003593_03884
You are here: Home > Sequence: MGYG000003593_03884
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
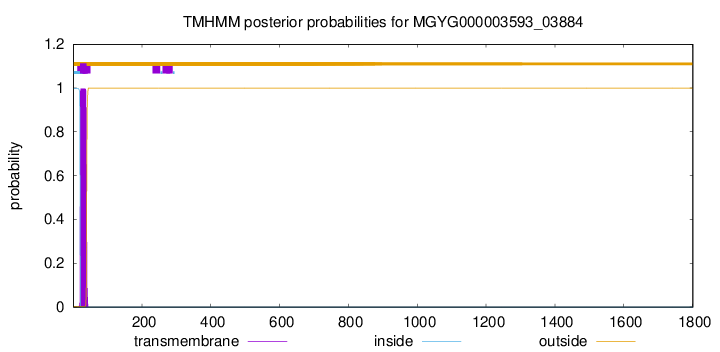
TMHMM annotations
Basic Information help
| Species | Enterocloster sp900770345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster sp900770345 | |||||||||||
| CAZyme ID | MGYG000003593_03884 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34014; End: 39419 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 1014 | 1284 | 1.5e-49 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 4.63e-47 | 1004 | 1392 | 43 | 354 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 1.16e-24 | 1013 | 1626 | 96 | 648 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 7.71e-22 | 1004 | 1284 | 49 | 282 | Glycosyl hydrolase family 3 N terminal domain. |
| COG5263 | COG5263 | 1.75e-13 | 1694 | 1800 | 220 | 313 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033930 | pneumo_PspA | 2.45e-13 | 1681 | 1769 | 438 | 514 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARE65206.1 | 0.0 | 849 | 1799 | 17 | 923 |
| QQR04093.1 | 0.0 | 849 | 1799 | 17 | 923 |
| QIX92882.1 | 6.88e-258 | 857 | 1624 | 8 | 744 |
| ASN94235.1 | 5.28e-253 | 857 | 1624 | 8 | 744 |
| QRP41088.1 | 5.28e-253 | 857 | 1624 | 8 | 744 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M6G_A | 3.37e-25 | 1019 | 1577 | 120 | 598 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 6R5I_A | 2.88e-24 | 1050 | 1634 | 88 | 624 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5R_A | 1.15e-23 | 1050 | 1634 | 89 | 625 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5O_A | 1.15e-23 | 1050 | 1634 | 88 | 624 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5O_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5R_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5P_A | 1.24e-23 | 1050 | 1634 | 120 | 656 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5P_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5BCC6 | 3.49e-59 | 894 | 1626 | 37 | 615 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| B8NGU6 | 3.85e-58 | 894 | 1580 | 41 | 605 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q2UFP8 | 6.07e-57 | 883 | 1580 | 34 | 609 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q46684 | 1.17e-55 | 875 | 1401 | 38 | 501 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| T2KMH9 | 4.65e-25 | 1013 | 1626 | 113 | 638 | Putative beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22230 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
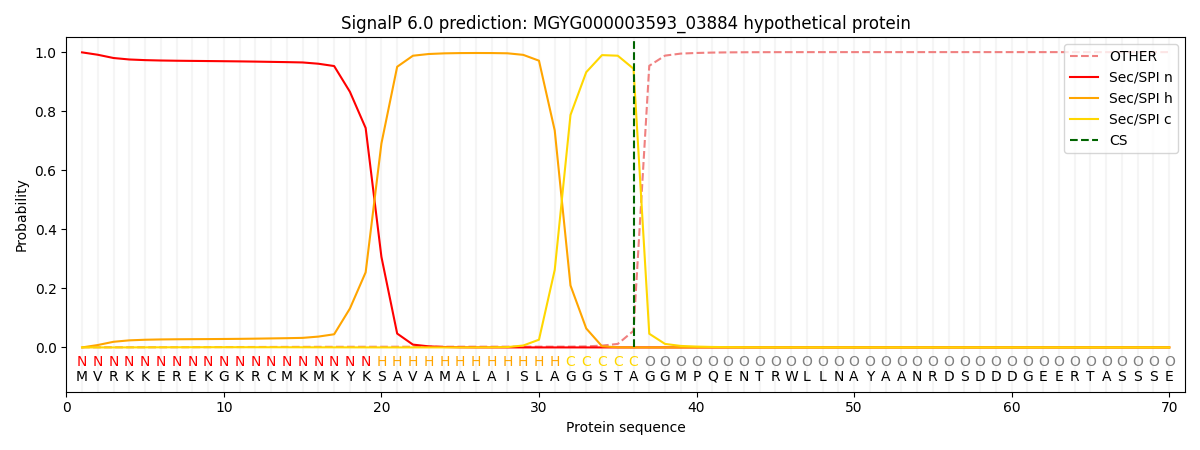
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001303 | 0.997051 | 0.000997 | 0.000218 | 0.000202 | 0.000192 |