You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003615_01723
You are here: Home > Sequence: MGYG000003615_01723
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-791 sp900315055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-791; CAG-791 sp900315055 | |||||||||||
| CAZyme ID | MGYG000003615_01723 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37607; End: 41257 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 34 | 430 | 2e-74 | 0.9628378378378378 |
| GH18 | 862 | 1086 | 1.6e-23 | 0.652027027027027 |
| CBM5 | 654 | 694 | 9.4e-17 | 0.95 |
| CBM5 | 744 | 784 | 4.2e-16 | 0.95 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06548 | GH18_chitinase | 5.12e-110 | 36 | 422 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| COG3325 | ChiA | 1.88e-103 | 12 | 431 | 17 | 432 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
| cd02871 | GH18_chitinase_D-like | 5.41e-91 | 861 | 1147 | 1 | 308 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| smart00636 | Glyco_18 | 5.12e-89 | 35 | 422 | 1 | 334 | Glyco_18 domain. |
| pfam00704 | Glyco_hydro_18 | 3.78e-76 | 35 | 422 | 1 | 307 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXF14593.1 | 1.92e-213 | 38 | 578 | 1 | 543 |
| QXF14594.1 | 3.84e-161 | 97 | 578 | 1 | 470 |
| CBH26257.1 | 4.01e-152 | 10 | 694 | 19 | 757 |
| QDA73531.1 | 1.54e-151 | 10 | 694 | 19 | 757 |
| QPJ26701.1 | 4.24e-151 | 10 | 694 | 19 | 757 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EBV_A | 5.82e-77 | 861 | 1148 | 5 | 287 | Crystalstructure of putative Chitinase A from Streptomyces coelicolor. [Streptomyces coelicolor] |
| 6KST_A | 2.54e-58 | 29 | 435 | 2 | 370 | Crystalstructure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KST_B Crystal structure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KXL_A Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXL_B Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 6KXM_A | 1.18e-57 | 29 | 435 | 2 | 370 | Crystalstructure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXM_B Crystal structure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 1ITX_A | 1.37e-56 | 23 | 435 | 1 | 419 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
| 6KXN_A | 1.87e-56 | 29 | 435 | 2 | 370 | Crystalstructure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXN_B Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 1.89e-53 | 23 | 435 | 33 | 451 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P27050 | 3.18e-53 | 856 | 1159 | 185 | 518 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| P32823 | 4.67e-53 | 29 | 697 | 153 | 818 | Chitinase A OS=Pseudoalteromonas piscicida OX=43662 GN=chiA PE=1 SV=1 |
| P32470 | 2.11e-48 | 14 | 420 | 18 | 381 | Chitinase 1 OS=Aphanocladium album OX=12942 GN=CHI1 PE=1 SV=2 |
| P07254 | 8.02e-48 | 39 | 439 | 163 | 560 | Chitinase A OS=Serratia marcescens OX=615 GN=chiA PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
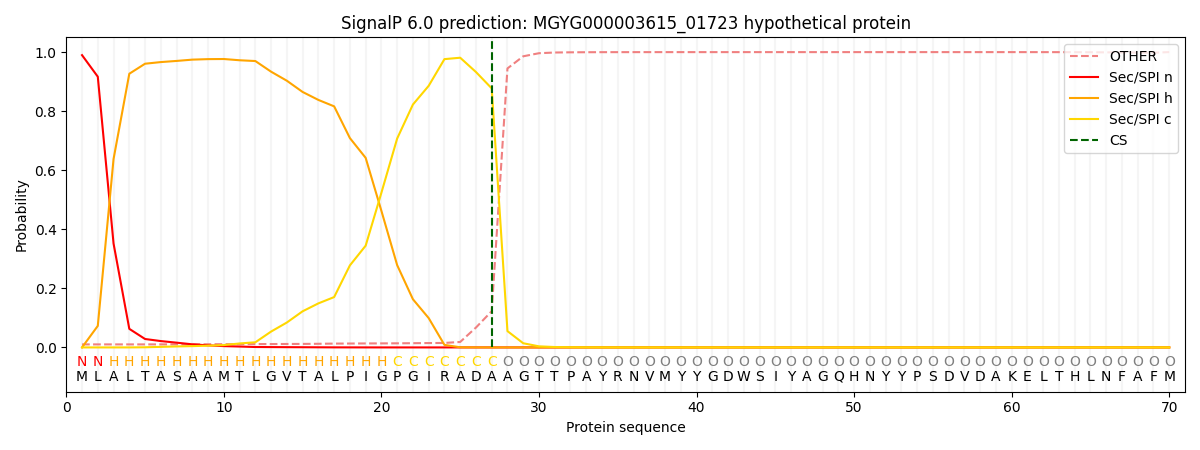
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014487 | 0.983773 | 0.000304 | 0.000626 | 0.000406 | 0.000358 |
