You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003617_01679
You are here: Home > Sequence: MGYG000003617_01679
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Helicobacter_D sp900770765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Helicobacteraceae; Helicobacter_D; Helicobacter_D sp900770765 | |||||||||||
| CAZyme ID | MGYG000003617_01679 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase C | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6664; End: 8013 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 272 | 423 | 7.1e-23 | 0.8074074074074075 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK11671 | mltC | 2.97e-111 | 114 | 422 | 52 | 356 | membrane-bound lytic murein transglycosylase MltC. |
| cd16893 | LT_MltC_MltE | 1.78e-91 | 259 | 420 | 1 | 162 | membrane-bound lytic murein transglycosylases MltC and MltE, and similar proteins. MltC and MltE are periplasmic, outer membrane attached lytic transglycosylases (LTs), which cleave beta-1,4-glycosidic bonds joining N-acetylmuramic acid and N-acetylglucosamine in the cell wall peptidoglycan, yielding 1,6-anhydromuropeptides. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda |
| pfam11873 | DUF3393 | 1.68e-66 | 99 | 256 | 8 | 161 | Domain of unknown function (DUF3393). This domain is functionally uncharacterized. This domain is found in bacteria. This presumed domain is typically between 188 to 206 amino acids in length. This domain is found associated with pfam01464. |
| cd16896 | LT_Slt70-like | 1.52e-33 | 256 | 372 | 3 | 109 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG0741 | MltE | 1.65e-31 | 133 | 418 | 17 | 279 | Soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOQ98835.1 | 2.50e-186 | 73 | 423 | 22 | 372 |
| VEJ06639.1 | 4.47e-180 | 1 | 436 | 1 | 415 |
| AWI35046.1 | 2.59e-176 | 83 | 423 | 51 | 391 |
| VEG80404.1 | 5.25e-122 | 86 | 422 | 53 | 386 |
| CAE10356.1 | 5.25e-122 | 86 | 422 | 53 | 386 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4C5F_A | 4.21e-77 | 99 | 423 | 19 | 339 | Structureof Lytic Transglycosylase MltC from Escherichia coli at 2.3 A resolution. [Escherichia coli],4C5F_B Structure of Lytic Transglycosylase MltC from Escherichia coli at 2.3 A resolution. [Escherichia coli] |
| 4CFO_A | 1.18e-76 | 99 | 423 | 19 | 339 | Structureof Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. [Escherichia coli],4CFO_B Structure of Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. [Escherichia coli],4CFP_A Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution [Escherichia coli],4CFP_B Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution [Escherichia coli],4CHX_A Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 [Escherichia coli],4CHX_B Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 [Escherichia coli] |
| 6GI4_B | 2.19e-22 | 263 | 418 | 28 | 182 | Structureof Lytic Transglycosylase MltE mutant S75A from E.coli [Escherichia coli] |
| 2Y8P_A | 3.00e-22 | 263 | 418 | 28 | 182 | CrystalStructure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli [Escherichia coli K-12],2Y8P_B Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli [Escherichia coli K-12] |
| 3T36_A | 3.71e-22 | 263 | 418 | 45 | 199 | Crystalstructure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_B Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_C Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_D Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],3T36_E Crystal structure of lytic transglycosylase MltE from Eschericha coli [Escherichia coli K-12],4HJV_A Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_B Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_C Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_D Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12],4HJV_E Crystal structure of E. coli MltE with bound bulgecin and murodipeptide [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9R6R2 | 1.27e-80 | 89 | 423 | 27 | 356 | Membrane-bound lytic murein transglycosylase C OS=Yersinia pestis bv. Antiqua (strain Angola) OX=349746 GN=mltC PE=3 SV=1 |
| Q1CB94 | 1.27e-80 | 89 | 423 | 27 | 356 | Membrane-bound lytic murein transglycosylase C OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=mltC PE=3 SV=2 |
| Q1CEV1 | 1.27e-80 | 89 | 423 | 27 | 356 | Membrane-bound lytic murein transglycosylase C OS=Yersinia pestis bv. Antiqua (strain Nepal516) OX=377628 GN=mltC PE=3 SV=2 |
| B2K0V2 | 1.27e-80 | 89 | 423 | 27 | 356 | Membrane-bound lytic murein transglycosylase C OS=Yersinia pseudotuberculosis serotype IB (strain PB1/+) OX=502801 GN=mltC PE=3 SV=1 |
| A7FEX3 | 1.27e-80 | 89 | 423 | 27 | 356 | Membrane-bound lytic murein transglycosylase C OS=Yersinia pseudotuberculosis serotype O:1b (strain IP 31758) OX=349747 GN=mltC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
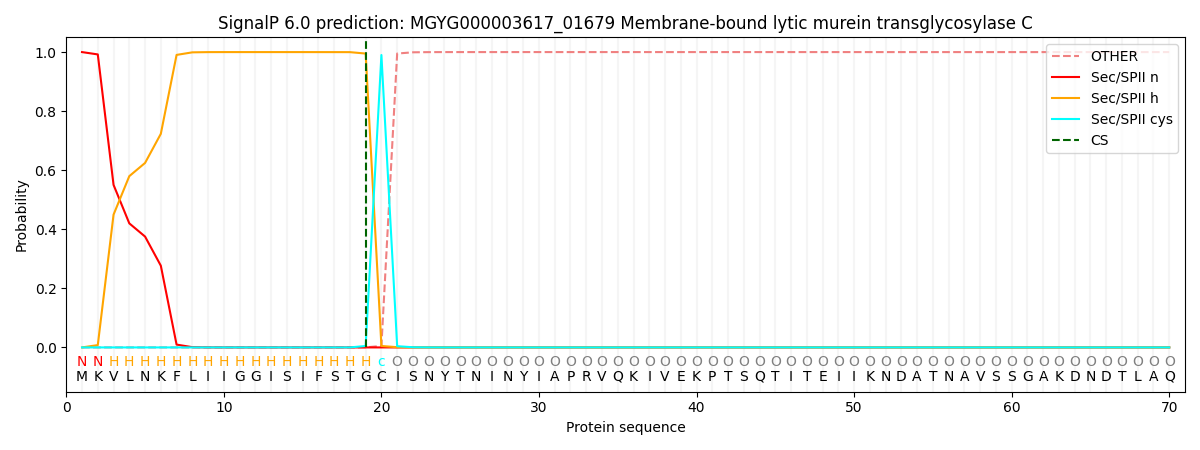
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000046 | 0.000000 | 0.000000 | 0.000000 |
