You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003626_02128
You are here: Home > Sequence: MGYG000003626_02128
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900771005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900771005 | |||||||||||
| CAZyme ID | MGYG000003626_02128 | |||||||||||
| CAZy Family | GH146 | |||||||||||
| CAZyme Description | Hydroxyacylglutathione hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18279; End: 21431 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 36 | 540 | 5.1e-202 | 0.9980119284294234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 3.72e-168 | 36 | 540 | 1 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 4.77e-115 | 37 | 547 | 12 | 508 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| cd07712 | MBLAC2-like_MBL-fold | 1.54e-46 | 829 | 991 | 10 | 182 | uncharacterized human metallo-beta-lactamase domain-containing protein 2 and related proteins; MBL-fold metallo hydrolase domain. Includes the MBL-fold metallo hydrolase domain of uncharacterized human MBLAC2 and related proteins. Members of this subgroup belong to the MBL-fold metallo-hydrolase superfamily which is comprised mainly of hydrolytic enzymes which carry out a variety of biological functions. |
| pfam16375 | DUF4986 | 1.77e-35 | 546 | 630 | 3 | 84 | Domain of unknown function. This family around 150 residues locates in the C-terminal of some uncharacterized proteins in various Bacteroides and Bacillus species. The function of this family remains unknown. |
| cd06262 | metallo-hydrolase-like_MBL-fold | 6.60e-23 | 825 | 991 | 7 | 188 | mainly hydrolytic enzymes and related proteins which carry out various biological functions; MBL-fold metallohydrolase domain. Members of the MBL-fold metallohydrolase superfamily are mainly hydrolytic enzymes which carry out a variety of biological functions. The class B metal beta-lactamases (MBLs) for which this fold was named perform only a small fraction of the activities included in this superfamily. Activities carried out by superfamily members include class B beta-lactamases which can catalyze the hydrolysis of a wide range of beta-lactam antibiotics, hydroxyacylglutathione hydrolases (also called glyoxalase II) which hydrolyze S-d-lactoylglutathione to d-lactate in the second step of the glycoxlase system, AHL lactonases which catalyze the hydrolysis and opening of the homoserine lactone rings of acyl homoserine lactones (AHLs), persulfide dioxygenase which catalyze the oxidation of glutathione persulfide to glutathione and persulfite in the mitochondria, flavodiiron proteins which catalyze the reduction of oxygen and/or nitric oxide to water or nitrous oxide respectively, cleavage and polyadenylation specificity factors such as the Int9 and Int11 subunits of Integrator, Sdsa1-like and AtsA-like arylsulfatases, 5'-exonucleases human SNM1A and yeast Pso2p, ribonuclease J which has both 5'-3' exoribonucleolytic and endonucleolytic activity and ribonuclease Z which catalyzes the endonucleolytic removal of the 3' extension of the majority of tRNA precursors, cyclic nucleotide phosphodiesterases which decompose cyclic adenosine and guanosine 3', 5'-monophosphate (cAMP and cGMP) respectively, insecticide hydrolases, and proteins required for natural transformation competence. The diversity of biological roles is reflected in variations in the active site metallo-chemistry, for example classical members of the superfamily are di-, or less commonly mono-, zinc-ion-dependent hydrolases, human persulfide dioxygenase ETHE1 is a mono-iron binding member of the superfamily; Arabidopsis thaliana hydroxyacylglutathione hydrolases incorporates iron, manganese, and zinc in its dinuclear metal binding site, and flavodiiron proteins contains a diiron site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD36520.1 | 0.0 | 11 | 792 | 9 | 786 |
| ADY37333.1 | 0.0 | 1 | 793 | 1 | 797 |
| QCD43163.1 | 2.28e-310 | 20 | 792 | 16 | 785 |
| QUT88913.1 | 3.05e-304 | 13 | 791 | 10 | 796 |
| SCV06553.1 | 3.75e-304 | 22 | 793 | 22 | 798 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YQH_AAA | 4.17e-224 | 23 | 792 | 19 | 802 | ChainAAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
| 5OPJ_A | 1.44e-215 | 23 | 792 | 19 | 802 | Beta-L-arabinofuranosidase[Bacteroides thetaiotaomicron] |
| 6EX6_A | 1.37e-21 | 125 | 542 | 106 | 547 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 4QJY_A | 1.18e-16 | 82 | 538 | 131 | 558 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 4QK0_A | 1.42e-15 | 82 | 538 | 131 | 558 | Crystalstructure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5F336 | 1.03e-16 | 829 | 1003 | 31 | 240 | Acyl-coenzyme A thioesterase MBLAC2 OS=Gallus gallus OX=9031 GN=MBLAC2 PE=2 SV=1 |
| A5PJT0 | 3.55e-16 | 829 | 1003 | 31 | 243 | Acyl-coenzyme A thioesterase MBLAC2 OS=Bos taurus OX=9913 GN=MBLAC2 PE=2 SV=1 |
| Q68D91 | 3.55e-16 | 829 | 1003 | 31 | 243 | Acyl-coenzyme A thioesterase MBLAC2 OS=Homo sapiens OX=9606 GN=MBLAC2 PE=1 SV=3 |
| Q8BL86 | 3.55e-16 | 829 | 1003 | 31 | 243 | Acyl-coenzyme A thioesterase MBLAC2 OS=Mus musculus OX=10090 GN=Mblac2 PE=1 SV=2 |
| Q5XGR8 | 2.09e-07 | 831 | 991 | 35 | 198 | Endoribonuclease LACTB2 OS=Xenopus laevis OX=8355 GN=lactb2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
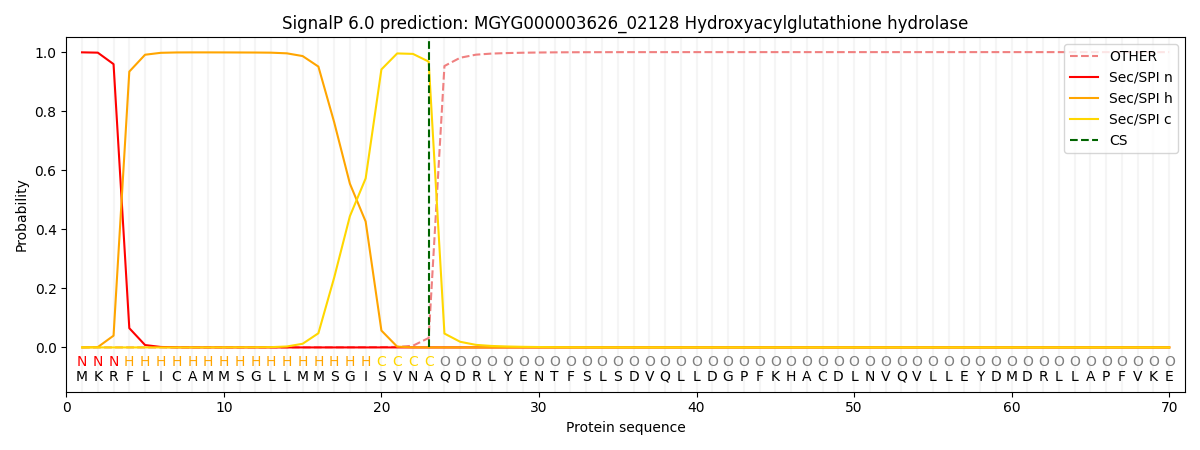
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000563 | 0.997592 | 0.001193 | 0.000252 | 0.000188 | 0.000163 |
