You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003627_01517
You are here: Home > Sequence: MGYG000003627_01517
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1732; | |||||||||||
| CAZyme ID | MGYG000003627_01517 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 728; End: 1840 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 36 | 360 | 1.1e-104 | 0.9834983498349835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 2.78e-116 | 34 | 360 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.47e-113 | 75 | 358 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.91e-98 | 12 | 366 | 5 | 345 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| cd17542 | REC_CheY | 0.008 | 188 | 266 | 37 | 100 | phosphoacceptor receiver (REC) domain of chemotaxis protein CheY. The chemotaxis response regulator CheY contains a stand-alone REC domain. Chemotaxis is a behavior known for motile bacteria that directs their movement in response to chemical gradients. CheY is involved in transmitting sensory signals from chemoreceptors to the flagellar motors. Phosphorylated CheY interacts with the flagella switch components FliM and FliY, which causes counterclockwise rotation of the flagella, resulting in smooth swimming. REC domains function as phosphorylation-mediated switches within response regulators, but some also transfer phosphoryl groups in multistep phosphorelays. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFN75725.1 | 8.82e-98 | 11 | 363 | 3 | 369 |
| ACW02028.1 | 4.57e-91 | 42 | 359 | 6 | 338 |
| ACP87400.1 | 4.57e-91 | 42 | 359 | 6 | 338 |
| ACW01955.1 | 4.51e-90 | 10 | 361 | 7 | 372 |
| ACP87327.1 | 4.51e-90 | 10 | 361 | 7 | 372 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 7.09e-81 | 28 | 359 | 7 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 2Q8X_A | 3.13e-80 | 33 | 361 | 7 | 331 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 1N82_A | 1.76e-79 | 33 | 361 | 7 | 331 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 3MS8_A | 2.49e-79 | 33 | 361 | 7 | 331 | Enzyme-Substrateinteractions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MS8_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 3MUI_A | 1.40e-78 | 33 | 361 | 7 | 331 | Enzyme-Substrateinteractions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUI_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48789 | 1.91e-79 | 1 | 361 | 1 | 367 | Endo-1,4-beta-xylanase A OS=Prevotella ruminicola OX=839 GN=xynA PE=3 SV=1 |
| O69231 | 1.58e-77 | 42 | 359 | 16 | 329 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| P49942 | 1.03e-74 | 11 | 365 | 8 | 375 | Endo-1,4-beta-xylanase A OS=Bacteroides ovatus OX=28116 GN=xylI PE=2 SV=1 |
| P45703 | 1.87e-72 | 33 | 361 | 7 | 330 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=1 SV=1 |
| P23556 | 3.77e-72 | 31 | 359 | 18 | 340 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
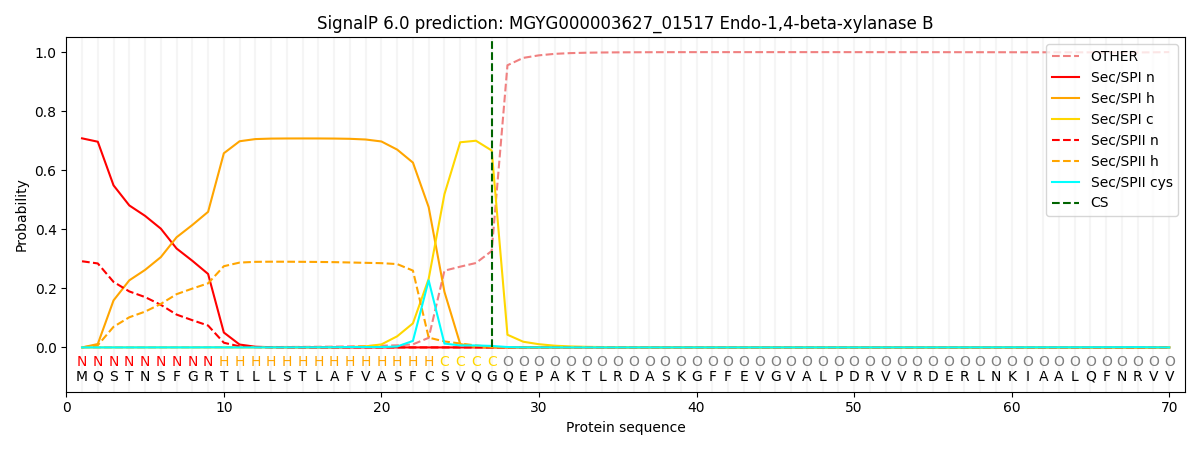
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000813 | 0.701028 | 0.297421 | 0.000282 | 0.000239 | 0.000197 |
