You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003633_00535
You are here: Home > Sequence: MGYG000003633_00535
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Paludibacteraceae; RF16; | |||||||||||
| CAZyme ID | MGYG000003633_00535 | |||||||||||
| CAZy Family | CBM6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1200; End: 3584 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11576 | GH99_GH71_like_2 | 1.65e-142 | 183 | 576 | 1 | 378 | Uncharacterized glycoside hydrolase family 99-like domain. This family of putative glycoside hydrolases resembles glycosyl hydrolase families 71 and 99 (following the CAZY nomenclature) and may share a similar catalytic site and mechanism. The domain may co-occur with other domains involved in the binding/processing of glycans. |
| cd11573 | GH99_GH71_like | 1.53e-24 | 260 | 536 | 5 | 256 | Glycoside hydrolase families 71, 99, and related domains. This superfamily of glycoside hydrolases contains families GH71 and GH99 (following the CAZY nomenclature), as well as other members with undefined function and specificity. |
| pfam09044 | Kp4 | 0.008 | 374 | 411 | 76 | 111 | Kp4. Members of this fungal family of toxins specifically inhibit voltage-gated calcium channels in mammalian cells. They adopt an alpha/beta-sandwich structure, comprising a five-stranded antiparallel beta-sheet with two antiparallel alpha-helices lying at approximately 45 degrees to these strands. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLG40767.1 | 4.04e-103 | 182 | 584 | 170 | 559 |
| QOS78953.1 | 6.40e-101 | 182 | 584 | 170 | 559 |
| QGG55654.1 | 4.49e-100 | 180 | 572 | 226 | 606 |
| ASS66155.1 | 9.52e-100 | 182 | 584 | 170 | 559 |
| BBI31685.1 | 3.06e-99 | 129 | 584 | 259 | 693 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
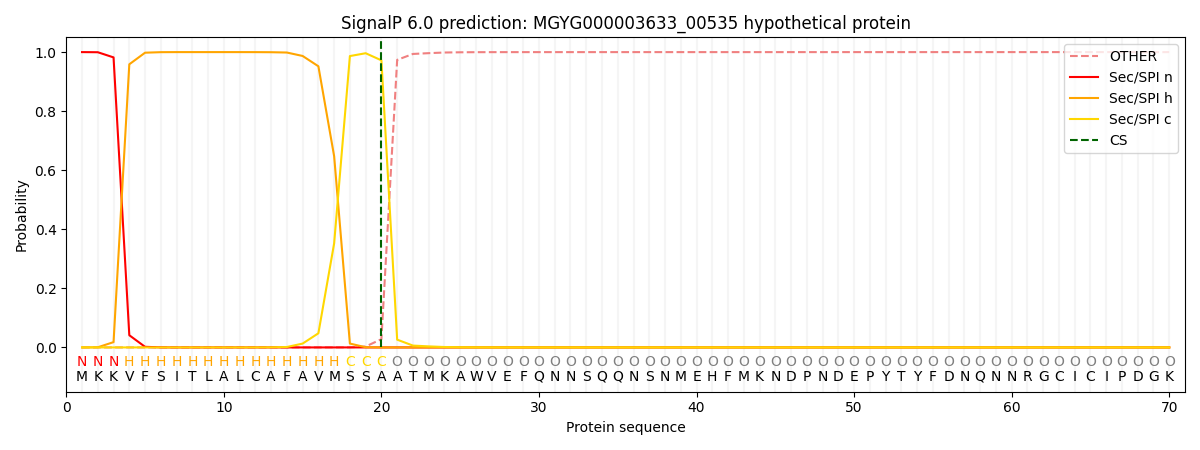
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000398 | 0.998850 | 0.000223 | 0.000178 | 0.000171 | 0.000170 |
