You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003637_01359
You are here: Home > Sequence: MGYG000003637_01359
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | W0P29-029 sp004558035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; UBA953; W0P29-029; W0P29-029 sp004558035 | |||||||||||
| CAZyme ID | MGYG000003637_01359 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3793; End: 6159 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 201 | 756 | 1.2e-174 | 0.9908759124087592 |
| CBM51 | 48 | 182 | 3.7e-27 | 0.9328358208955224 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 1.71e-28 | 43 | 182 | 5 | 134 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 4.25e-24 | 41 | 182 | 5 | 143 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam13229 | Beta_helix | 1.38e-05 | 598 | 757 | 31 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 4.99e-05 | 598 | 736 | 8 | 117 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05048 | NosD | 0.008 | 596 | 667 | 91 | 152 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWO89927.1 | 8.80e-267 | 41 | 776 | 37 | 750 |
| QWO84961.1 | 8.80e-267 | 41 | 776 | 37 | 750 |
| QWO94795.1 | 8.80e-267 | 41 | 776 | 37 | 750 |
| QWP69603.1 | 8.80e-267 | 41 | 776 | 37 | 750 |
| QWO92264.1 | 8.80e-267 | 41 | 776 | 37 | 750 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 2.48e-72 | 198 | 727 | 28 | 564 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 1.27e-71 | 198 | 727 | 28 | 564 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2UNU8 | 4.23e-263 | 29 | 787 | 57 | 793 | Alpha-1,3-galactosidase B OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaB PE=3 SV=1 |
| A6LFT2 | 1.62e-111 | 178 | 756 | 6 | 591 | Alpha-1,3-galactosidase B OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=glaB PE=3 SV=1 |
| A6L2M8 | 2.15e-109 | 198 | 756 | 24 | 593 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB2 PE=3 SV=1 |
| A6KWM0 | 6.61e-107 | 202 | 753 | 28 | 590 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB1 PE=3 SV=1 |
| Q5LGZ8 | 8.18e-105 | 199 | 753 | 28 | 593 | Alpha-1,3-galactosidase B OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
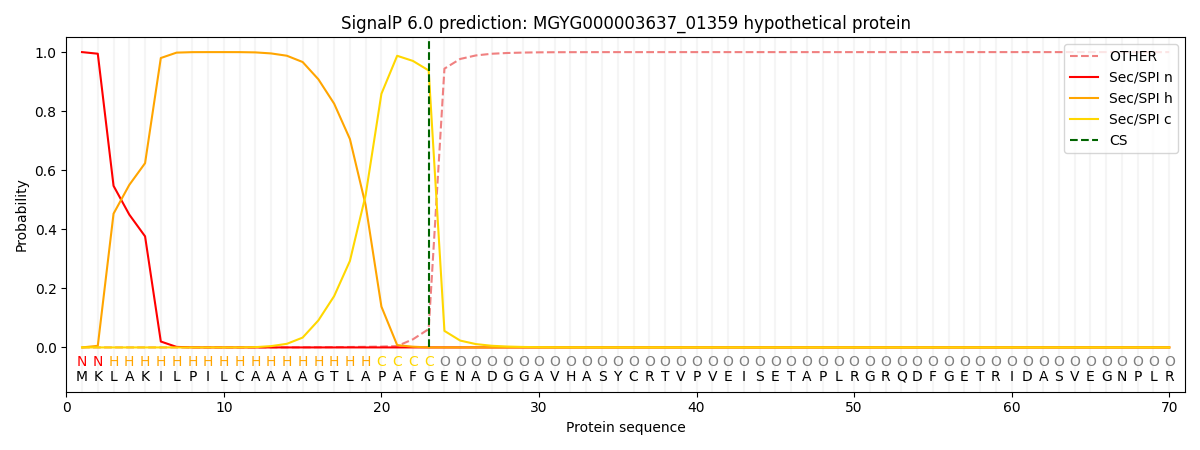
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000466 | 0.998726 | 0.000270 | 0.000191 | 0.000166 | 0.000161 |
