You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003642_01263
You are here: Home > Sequence: MGYG000003642_01263
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
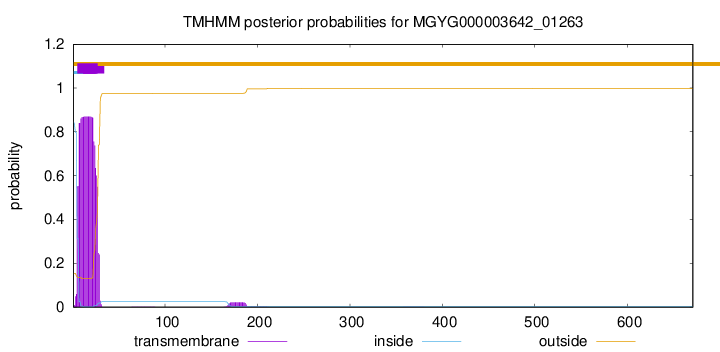
TMHMM annotations
Basic Information help
| Species | RUG572 sp900771305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900771305 | |||||||||||
| CAZyme ID | MGYG000003642_01263 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2157; End: 4172 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 298 | 655 | 4e-66 | 0.9107692307692308 |
| CE1 | 65 | 258 | 1.4e-25 | 0.8722466960352423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 8.24e-69 | 287 | 588 | 100 | 399 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| COG4099 | COG4099 | 2.70e-40 | 57 | 278 | 168 | 387 | Predicted peptidase [General function prediction only]. |
| pfam00295 | Glyco_hydro_28 | 6.19e-16 | 418 | 623 | 76 | 273 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| COG3509 | LpqC | 1.25e-11 | 43 | 231 | 27 | 207 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG0412 | DLH | 1.91e-10 | 166 | 257 | 109 | 201 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQY41723.1 | 1.86e-153 | 288 | 668 | 71 | 450 |
| ALK86407.1 | 1.51e-152 | 293 | 668 | 41 | 415 |
| QUT56858.1 | 4.82e-152 | 293 | 668 | 75 | 449 |
| ABR37881.1 | 4.82e-152 | 293 | 668 | 75 | 449 |
| QQY43094.1 | 4.82e-152 | 293 | 668 | 75 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 9.57e-142 | 283 | 668 | 58 | 442 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3JUR_A | 7.08e-68 | 288 | 660 | 46 | 417 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 3DOH_A | 2.52e-45 | 58 | 278 | 153 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 4.84e-31 | 60 | 278 | 18 | 217 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 4Q82_A | 1.16e-16 | 80 | 278 | 81 | 277 | CrystalStructure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365],4Q82_B Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27644 | 1.32e-16 | 435 | 582 | 30 | 181 | Polygalacturonase OS=Rhizobium radiobacter OX=358 GN=pgl PE=2 SV=1 |
| Q9LW07 | 8.59e-09 | 426 | 552 | 137 | 257 | Probable polygalacturonase At3g15720 OS=Arabidopsis thaliana OX=3702 GN=At3g15720 PE=3 SV=1 |
| Q94AJ5 | 1.27e-06 | 313 | 587 | 103 | 330 | Probable polygalacturonase At1g80170 OS=Arabidopsis thaliana OX=3702 GN=At1g80170 PE=1 SV=1 |
| Q5AYH4 | 5.71e-06 | 321 | 651 | 202 | 492 | Probable endopolygalacturonase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pgaD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
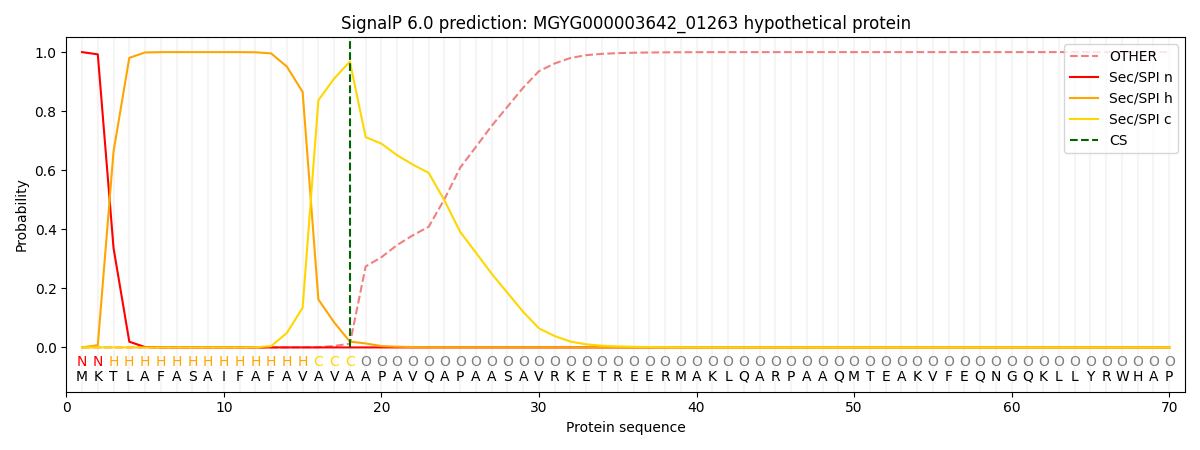
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000336 | 0.998928 | 0.000180 | 0.000190 | 0.000175 | 0.000164 |