You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003642_01338
Basic Information
help
| Species |
RUG572 sp900771305
|
| Lineage |
Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900771305
|
| CAZyme ID |
MGYG000003642_01338
|
| CAZy Family |
GH39 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 630 |
|
69387.78 |
6.8224 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003642 |
3810265 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 7119;
End: 9011
Strand: +
|
No EC number prediction in MGYG000003642_01338.
| Family |
Start |
End |
Evalue |
family coverage |
| GH39 |
63 |
238 |
1.8e-29 |
0.4965197215777262 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam01229
|
Glyco_hydro_39 |
4.21e-08 |
67 |
231 |
74 |
270 |
Glycosyl hydrolases family 39. |
| COG3664
|
XynB |
1.89e-04 |
99 |
248 |
117 |
266 |
Beta-xylosidase [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4EKJ_A
|
4.76e-09 |
35 |
196 |
46 |
231 |
ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 4M29_A
|
4.76e-09 |
35 |
196 |
46 |
231 |
Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
This protein is predicted as OTHER
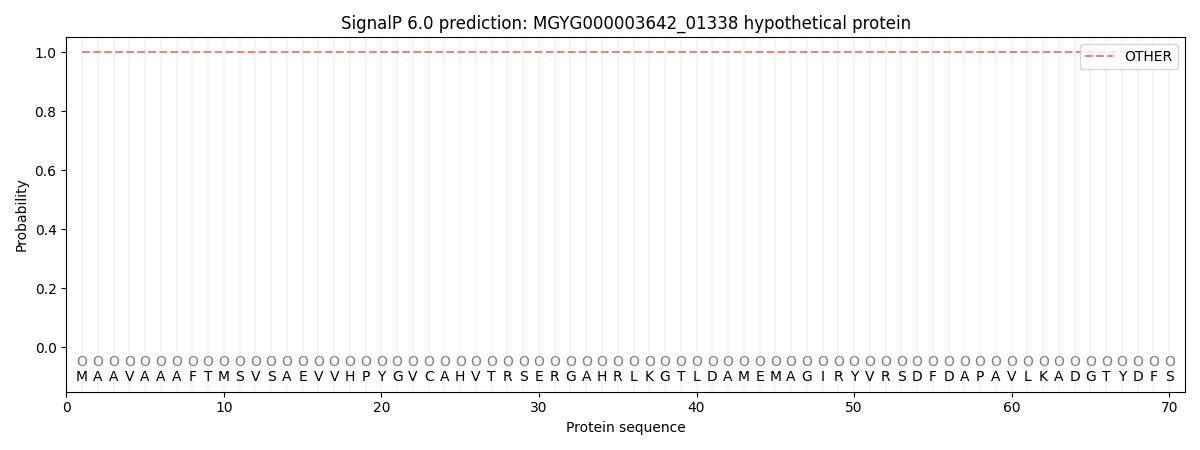
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000016
|
0.000022
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000003642_01338.

