You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003642_02040
Basic Information
help
| Species |
RUG572 sp900771305
|
| Lineage |
Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900771305
|
| CAZyme ID |
MGYG000003642_02040
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 672 |
|
75018.46 |
7.6592 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003642 |
3810265 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 6926;
End: 8944
Strand: +
|
No EC number prediction in MGYG000003642_02040.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
128 |
354 |
4.3e-34 |
0.8021978021978022 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG3934
|
COG3934 |
1.16e-14 |
46 |
558 |
6 |
511 |
Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
This protein is predicted as SP
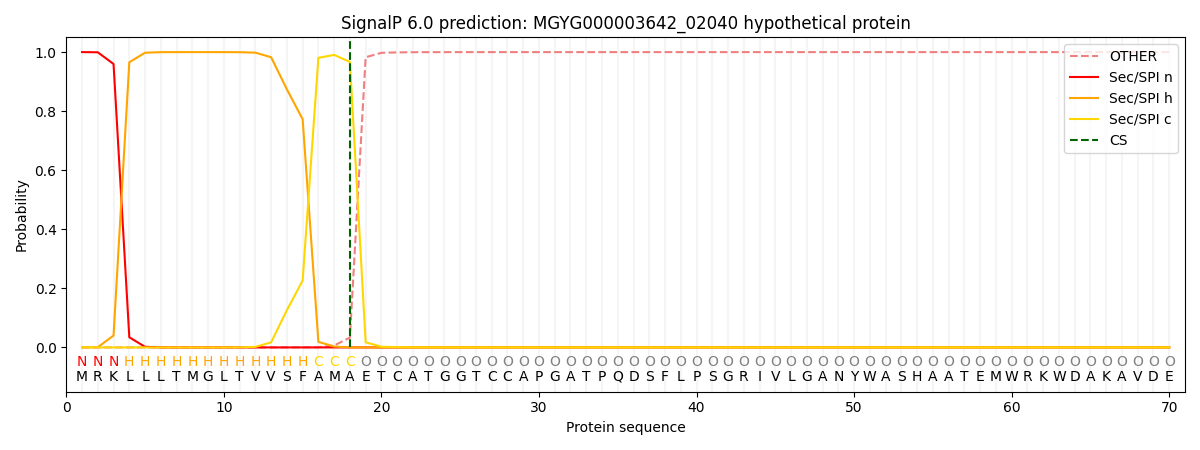
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000370
|
0.998810
|
0.000283
|
0.000174
|
0.000169
|
0.000160
|
There is no transmembrane helices in MGYG000003642_02040.

