You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003643_00067
You are here: Home > Sequence: MGYG000003643_00067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | W1P29-020 sp900771325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; W1P29-020; W1P29-020 sp900771325 | |||||||||||
| CAZyme ID | MGYG000003643_00067 | |||||||||||
| CAZy Family | CBM47 | |||||||||||
| CAZyme Description | Hercynine oxygenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8620; End: 12279 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM47 | 68 | 140 | 3.1e-18 | 0.5546875 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03781 | FGE-sulfatase | 3.38e-43 | 961 | 1217 | 6 | 259 | Sulfatase-modifying factor enzyme 1. This domain is found in eukaryotic proteins required for post-translational sulfatase modification (SUMF1). These proteins are associated with the rare disorder multiple sulfatase deficiency (MSD). The protein product of the SUMF1 gene is FGE, formylglycine (FGly),-generating enzyme, which is a sulfatase. Sulfatases are enzymes essential for degradation and remodelling of sulfate esters, and formylglycine (FGly), the key catalytic in the active site, is unique to sulfatases. FGE is localized to the endoplasmic reticulum (ER) and interacts with and modifies the unfolded form of newly synthesized sulfatases. FGE is a single-domain monomer with a surprising paucity of secondary structure that adopts a unique fold which is stabilized by two Ca2+ ions. The effect of all mutations found in MSD patients is explained by the FGE structure, providing a molecular basis for MSD. A redox-active disulfide bond is present in the active site of FGE. An oxidized cysteine residue, possibly cysteine sulfenic acid, has been detected that may allow formulation of a structure-based mechanism for FGly formation from cysteine residues in all sulfatases. In Mycobacteria and Treponema denticola this enzyme functions as an iron(II)-dependent oxidoreductase. |
| pfam18582 | HZS_alpha | 2.23e-39 | 639 | 729 | 1 | 98 | Hydrazine synthase alpha subunit middle domain. The crystal structure of hydrazine synthase multiprotein complex isolated from the anammox organism Kuenenia stuttgartiensis implies a two-step mechanism for hydrazine synthesis: a three-electron reduction of nitric oxide to hydroxylamine at the active site of the gamma-subunit and its subsequent condensation with ammonia, yielding hydrazine in the active centre of the alpha-subunit. The alpha-subunit consists of three domains: an N-terminal domain which includes a six-bladed beta-propeller, a middle domain binding a pentacoordinated c-type haem (haem alphaI) and a C-terminal domain which harbours a bis-histidine-coordinated c-type haem (haem alphaII). This entry represents the middle domain of subunit alpha of hydrazine synthase (HZS). |
| COG1262 | YfmG | 4.01e-39 | 961 | 1216 | 54 | 310 | Formylglycine-generating enzyme, required for sulfatase activity, contains SUMF1/FGE domain [Posttranslational modification, protein turnover, chaperones]. |
| smart00607 | FTP | 1.12e-06 | 51 | 131 | 1 | 85 | eel-Fucolectin Tachylectin-4 Pentaxrin-1 Domain. |
| pfam00754 | F5_F8_type_C | 9.99e-05 | 68 | 154 | 11 | 89 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74006.1 | 2.18e-244 | 229 | 1219 | 326 | 1330 |
| AQT69174.1 | 8.38e-61 | 213 | 879 | 139 | 768 |
| ASV76038.1 | 1.63e-59 | 60 | 889 | 40 | 932 |
| BBH25058.1 | 4.61e-31 | 962 | 1219 | 22 | 250 |
| QJW89271.1 | 1.68e-29 | 952 | 1216 | 16 | 254 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5HHA_A | 1.29e-21 | 961 | 1218 | 43 | 285 | Structureof PvdO from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5HHA_B Structure of PvdO from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 1Y4J_A | 3.29e-14 | 1033 | 1218 | 115 | 267 | Crystalstructure of the paralogue of the human formylglycine generating enzyme [Homo sapiens],1Y4J_B Crystal structure of the paralogue of the human formylglycine generating enzyme [Homo sapiens] |
| 3LE0_A | 1.83e-13 | 52 | 133 | 10 | 91 | ChainA, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEG_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEI_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEK_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis] |
| 3CQO_A | 5.35e-13 | 24 | 136 | 120 | 234 | Crystalstructure of a f-lectin (fucolectin) from morone saxatilis (striped bass) serum [Morone saxatilis],3CQO_B Crystal structure of a f-lectin (fucolectin) from morone saxatilis (striped bass) serum [Morone saxatilis],3CQO_C Crystal structure of a f-lectin (fucolectin) from morone saxatilis (striped bass) serum [Morone saxatilis] |
| 4GWI_A | 6.27e-13 | 52 | 133 | 10 | 91 | His62 mutant of the lectin binding domain of lectinolysin complexed with Lewis y [Streptococcus mitis],4GWJ_A His 62 mutant of the lectin binding domain of Lectinolysin complexed with Lewis b [Streptococcus mitis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q58CP2 | 1.25e-17 | 950 | 1214 | 20 | 289 | Inactive C-alpha-formylglycine-generating enzyme 2 OS=Bos taurus OX=9913 GN=SUMF2 PE=2 SV=1 |
| Q5RCR5 | 1.47e-15 | 950 | 1218 | 20 | 293 | Inactive C-alpha-formylglycine-generating enzyme 2 OS=Pongo abelii OX=9601 GN=SUMF2 PE=2 SV=1 |
| Q8NBJ7 | 3.60e-15 | 950 | 1218 | 20 | 293 | Inactive C-alpha-formylglycine-generating enzyme 2 OS=Homo sapiens OX=9606 GN=SUMF2 PE=1 SV=2 |
| Q8BPG6 | 2.35e-14 | 949 | 1214 | 27 | 296 | Inactive C-alpha-formylglycine-generating enzyme 2 OS=Mus musculus OX=10090 GN=Sumf2 PE=1 SV=2 |
| Q8R0F3 | 1.14e-11 | 961 | 1218 | 90 | 366 | Formylglycine-generating enzyme OS=Mus musculus OX=10090 GN=Sumf1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
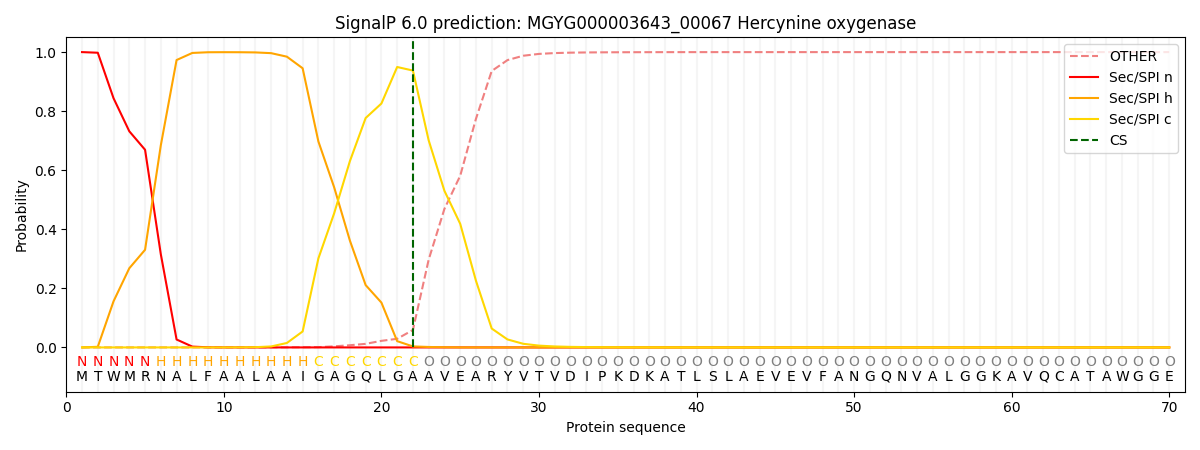
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000767 | 0.998346 | 0.000243 | 0.000233 | 0.000196 | 0.000202 |
