You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003648_01563
You are here: Home > Sequence: MGYG000003648_01563
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | NK4A136 sp002314855 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; NK4A136; NK4A136 sp002314855 | |||||||||||
| CAZyme ID | MGYG000003648_01563 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20228; End: 21496 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 96 | 366 | 2.8e-45 | 0.8415841584158416 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.16e-28 | 97 | 363 | 3 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 3.03e-25 | 118 | 363 | 67 | 305 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 7.64e-24 | 116 | 368 | 88 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARI46379.1 | 1.18e-194 | 9 | 407 | 5 | 386 |
| ARI46378.1 | 7.27e-185 | 1 | 408 | 1 | 390 |
| ARI46380.1 | 1.13e-181 | 9 | 410 | 9 | 402 |
| QTE68398.1 | 9.12e-177 | 9 | 421 | 6 | 405 |
| AHF24628.1 | 2.45e-174 | 9 | 422 | 6 | 406 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 1.13e-28 | 8 | 403 | 54 | 399 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 4.82e-27 | 8 | 403 | 54 | 399 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 2Q8X_A | 3.64e-20 | 101 | 347 | 53 | 301 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 1N82_A | 6.71e-20 | 101 | 347 | 53 | 301 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 3MS8_A | 2.28e-19 | 101 | 347 | 53 | 301 | Enzyme-Substrateinteractions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MS8_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY13 | 1.42e-20 | 98 | 360 | 77 | 358 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| A3DH97 | 3.59e-19 | 10 | 411 | 392 | 745 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| O69230 | 1.83e-17 | 101 | 402 | 419 | 741 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| P45703 | 5.27e-16 | 127 | 347 | 78 | 300 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=1 SV=1 |
| Q12603 | 2.89e-15 | 118 | 346 | 97 | 321 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
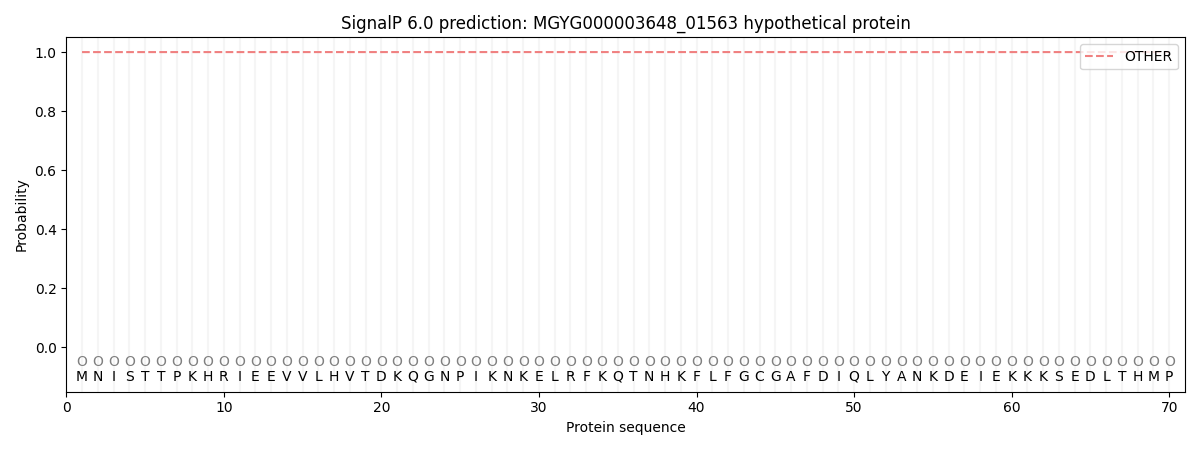
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000059 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
