You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003650_01355
You are here: Home > Sequence: MGYG000003650_01355
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1786 sp900771425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA1786; UBA1786 sp900771425 | |||||||||||
| CAZyme ID | MGYG000003650_01355 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 107698; End: 110217 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 19 | 835 | 6.3e-249 | 0.9939320388349514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 4.97e-39 | 143 | 614 | 392 | 866 | alpha-L-rhamnosidase. |
| pfam17132 | Glyco_hydro_106 | 5.54e-09 | 28 | 127 | 6 | 107 | alpha-L-rhamnosidase. |
| PRK10340 | ebgA | 7.67e-04 | 719 | 783 | 110 | 177 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALK86447.1 | 1.44e-305 | 1 | 835 | 1 | 914 |
| QQY43061.1 | 2.34e-304 | 1 | 835 | 1 | 914 |
| QUT56814.1 | 3.80e-303 | 1 | 835 | 1 | 914 |
| QQY39760.1 | 3.80e-303 | 1 | 835 | 1 | 914 |
| ABR37915.1 | 3.80e-303 | 1 | 835 | 1 | 914 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQM_A | 3.67e-36 | 144 | 839 | 412 | 1102 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 8.50e-36 | 144 | 839 | 412 | 1102 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6Q2F_A | 5.36e-24 | 143 | 835 | 455 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 2.95e-23 | 136 | 619 | 248 | 713 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
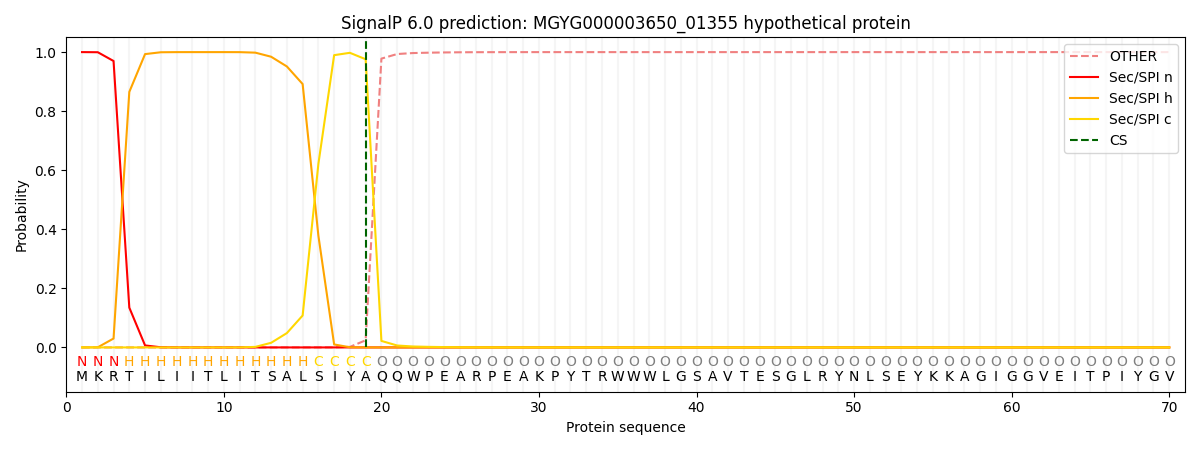
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000311 | 0.998904 | 0.000196 | 0.000198 | 0.000185 | 0.000165 |
