You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003650_01368
You are here: Home > Sequence: MGYG000003650_01368
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1786 sp900771425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA1786; UBA1786 sp900771425 | |||||||||||
| CAZyme ID | MGYG000003650_01368 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | Endoglucanase D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16914; End: 18365 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 32 | 480 | 8e-95 | 0.9808612440191388 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 6.88e-76 | 33 | 479 | 6 | 374 | Glycosyl hydrolase family 9. |
| PLN02345 | PLN02345 | 2.73e-10 | 84 | 298 | 38 | 244 | endoglucanase |
| PLN00119 | PLN00119 | 3.67e-10 | 84 | 482 | 73 | 488 | endoglucanase |
| PLN02613 | PLN02613 | 3.96e-10 | 81 | 275 | 65 | 253 | endoglucanase |
| PLN02909 | PLN02909 | 4.14e-09 | 84 | 455 | 76 | 431 | Endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADQ79913.1 | 4.21e-213 | 30 | 480 | 119 | 543 |
| QIA06920.1 | 2.60e-201 | 32 | 480 | 116 | 538 |
| AHW59138.1 | 1.21e-199 | 32 | 482 | 116 | 540 |
| QGY43879.1 | 4.01e-193 | 33 | 477 | 118 | 536 |
| BBE19007.1 | 6.11e-188 | 30 | 480 | 117 | 540 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CJ0_A | 3.32e-159 | 28 | 480 | 117 | 545 | ChainA, ENDOGLUCANASE D [Acetivibrio thermocellus],4CJ1_A Chain A, ENDOGLUCANASE D [Acetivibrio thermocellus] |
| 1CLC_A | 5.15e-159 | 28 | 480 | 131 | 559 | ChainA, ENDOGLUCANASE CELD; EC: 3.2.1.4 [Acetivibrio thermocellus] |
| 3X17_A | 2.36e-70 | 28 | 480 | 118 | 554 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
| 5U2O_A | 1.31e-56 | 31 | 478 | 87 | 535 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 6DHT_A | 4.31e-55 | 33 | 482 | 111 | 564 | Bacteroidesovatus GH9 Bacova_02649 [Bacteroides ovatus ATCC 8483] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C2S4 | 1.82e-158 | 28 | 480 | 117 | 545 | Endoglucanase D (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celD PE=1 SV=1 |
| A3DDN1 | 3.85e-158 | 28 | 480 | 141 | 569 | Endoglucanase D OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celD PE=1 SV=1 |
| A7LXT3 | 3.23e-54 | 33 | 482 | 125 | 578 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH9A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02649 PE=1 SV=1 |
| P23658 | 4.25e-52 | 18 | 483 | 72 | 546 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
| A3DCH1 | 4.73e-46 | 85 | 480 | 376 | 809 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celK PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
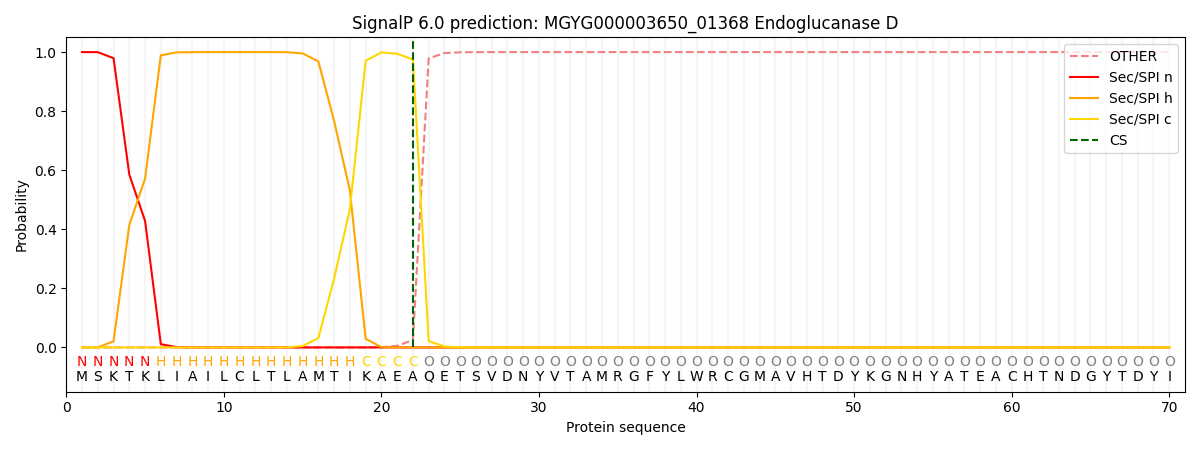
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000208 | 0.999173 | 0.000166 | 0.000159 | 0.000149 | 0.000138 |
