You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003651_00409
You are here: Home > Sequence: MGYG000003651_00409
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RF16 sp900767595 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Paludibacteraceae; RF16; RF16 sp900767595 | |||||||||||
| CAZyme ID | MGYG000003651_00409 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase C307 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17945; End: 19111 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 49 | 363 | 5.6e-150 | 0.9967948717948718 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.13e-22 | 65 | 363 | 20 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 9.10e-08 | 42 | 374 | 58 | 377 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd13676 | PBP2_TRAP_DctP2_like | 0.003 | 234 | 335 | 182 | 270 | Substrate-binding component of Tripartite ATP-independent Periplasmic transporter DctP2 and related proteins; the type 2 periplasmic-binding protein fold. This subgroup includes TRAP transporter DctP2 and its similar proteins. TRAP transporters are a large family of solute transporters ubiquitously found in bacteria and archaea. They are comprised of a periplasmic substrate-binding protein (SBP; often called the P subunit) and two unequally sized integral membrane components: a large transmembrane subunit involved in the translocation process (the M subunit) and a smaller membrane of unknown function (the Q subunit). The driving force of TRAP transporters is provided by electrochemical ion gradients (either protons or sodium ions) across the cytoplasmic membrane, rather than ATP hydrolysis. This substrate-binding domain belongs to the type 2 periplasmic binding fold protein superfamily (PBP2). The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. |
| cd13603 | PBP2_TRAP_Siap_TeaA_like | 0.009 | 234 | 384 | 182 | 297 | Substrate-binding domain of a sialic acid binding Tripartite ATP-independent Periplasmic transport system (SiaP) and related proteins; the type 2 periplasmic-binding protein fold. This subfamily includes the periplasmic-binding component of TRAP transport systems such as SiaP (a sialic acid binding virulence factor), TeaA (an ectoine binding protein), and an uncharacterized TM0322 from hyperthermophilic bacterium Thermotoga maritima. TRAP transporters are a large family of solute transporters ubiquitously found in bacteria and archaea. They are comprised of a periplasmic substrate-binding protein (SBP) and two unequally sized integral membrane components: a large transmembrane subunit involved in the translocation process and a smaller membrane of unknown function. The driving force of TRAP transporters is provided by electrochemical ion gradients (either protons or sodium ions) across the cytoplasmic membrane, rather than ATP hydrolysis. This substrate-binding domain belongs to the type 2 periplasmic binding fold protein superfamily (PBP2). The PBP2 proteins are typically comprised of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGK74985.1 | 6.40e-181 | 25 | 384 | 9 | 373 |
| ADB80112.1 | 8.54e-179 | 25 | 379 | 9 | 370 |
| AKF17200.1 | 5.32e-178 | 25 | 384 | 9 | 374 |
| ACA61171.1 | 6.14e-178 | 25 | 384 | 9 | 379 |
| ADE83057.1 | 5.27e-175 | 42 | 384 | 38 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CEC_A | 2.03e-36 | 49 | 383 | 7 | 338 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 1CEN_A | 5.47e-36 | 49 | 383 | 7 | 338 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
| 6KDD_A | 1.13e-26 | 41 | 374 | 15 | 309 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 3AMC_A | 3.45e-25 | 39 | 384 | 5 | 311 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
| 3AMG_A | 2.34e-24 | 39 | 384 | 5 | 311 | Crystalstructures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8],3AMG_B Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DJ77 | 1.53e-36 | 49 | 383 | 7 | 338 | Endoglucanase C OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celC PE=3 SV=1 |
| P23340 | 1.53e-36 | 49 | 383 | 7 | 338 | Endoglucanase C307 OS=Clostridium sp. (strain F1) OX=1508 GN=celC307 PE=1 SV=1 |
| P0C2S3 | 1.11e-35 | 49 | 383 | 7 | 338 | Endoglucanase C OS=Acetivibrio thermocellus OX=1515 GN=celC PE=1 SV=1 |
| W8QRE4 | 1.65e-06 | 30 | 366 | 4 | 342 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| P32603 | 8.62e-06 | 67 | 247 | 112 | 299 | Sporulation-specific glucan 1,3-beta-glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SPR1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
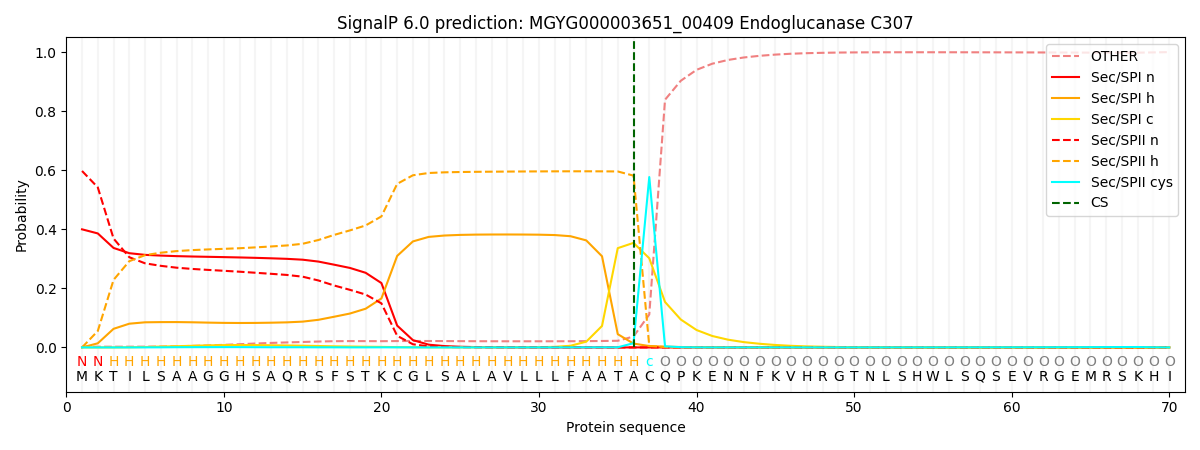
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003136 | 0.382762 | 0.613316 | 0.000207 | 0.000376 | 0.000193 |
