You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003658_00796
You are here: Home > Sequence: MGYG000003658_00796
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
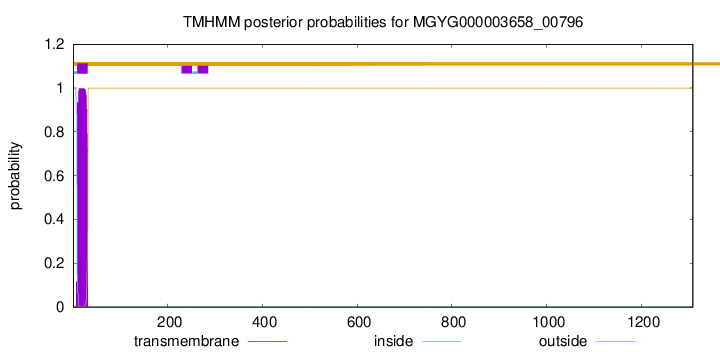
TMHMM annotations
Basic Information help
| Species | CAG-110 sp900546915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-110; CAG-110 sp900546915 | |||||||||||
| CAZyme ID | MGYG000003658_00796 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 185; End: 4114 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0737 | UshA | 3.24e-80 | 36 | 533 | 26 | 498 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09419 | PRK09419 | 2.08e-71 | 737 | 1305 | 40 | 602 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| PRK09419 | PRK09419 | 5.84e-70 | 29 | 559 | 653 | 1151 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| COG0737 | UshA | 9.70e-70 | 734 | 1259 | 22 | 517 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09558 | ushA | 2.58e-62 | 30 | 548 | 28 | 547 | bifunctional UDP-sugar hydrolase/5'-nucleotidase periplasmic precursor; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SET94084.1 | 7.77e-194 | 1 | 553 | 1 | 608 |
| ADL06114.1 | 3.05e-193 | 1 | 553 | 1 | 608 |
| QRV19771.1 | 3.05e-193 | 1 | 553 | 1 | 608 |
| ASN98642.1 | 7.80e-184 | 24 | 553 | 32 | 613 |
| QRP42178.1 | 7.80e-184 | 24 | 553 | 32 | 613 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2Z1A_A | 4.36e-40 | 29 | 557 | 22 | 534 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 4Q7F_A | 5.08e-36 | 739 | 1250 | 20 | 490 | ChainA, 5' nucleotidase family protein [Staphylococcus aureus subsp. aureus COL] |
| 3QFK_A | 5.08e-36 | 739 | 1250 | 20 | 490 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 4H2F_A | 1.00e-31 | 36 | 530 | 25 | 529 | Humanecto-5'-nucleotidase (CD73): crystal form I (open) in complex with adenosine [Homo sapiens],4H2G_A Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with adenosine [Homo sapiens] |
| 6XUQ_A | 1.13e-31 | 26 | 530 | 11 | 525 | HumanEcto-5'-nucleotidase (CD73) in complex with A1618 (compound 1b in publication) in the closed state in crystal form III [Homo sapiens],6Z9D_A Human Ecto-5'-nucleotidase (CD73) in complex with AOPCP derivative AB680 (compound 55 in publication) in the closed form (crystal form III) [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9BJC1 | 2.63e-49 | 8 | 508 | 3 | 460 | Mannosylglucosyl-3-phosphoglycerate phosphatase OS=Petrotoga mobilis (strain DSM 10674 / SJ95) OX=403833 GN=mggB PE=1 SV=1 |
| O34313 | 2.02e-45 | 36 | 557 | 668 | 1180 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| P44764 | 3.53e-42 | 712 | 1303 | 3 | 603 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=cpdB PE=3 SV=1 |
| P08331 | 5.76e-41 | 737 | 1305 | 22 | 595 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Escherichia coli (strain K12) OX=83333 GN=cpdB PE=1 SV=2 |
| P53052 | 3.60e-34 | 737 | 1305 | 27 | 600 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Yersinia enterocolitica OX=630 GN=cpdB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
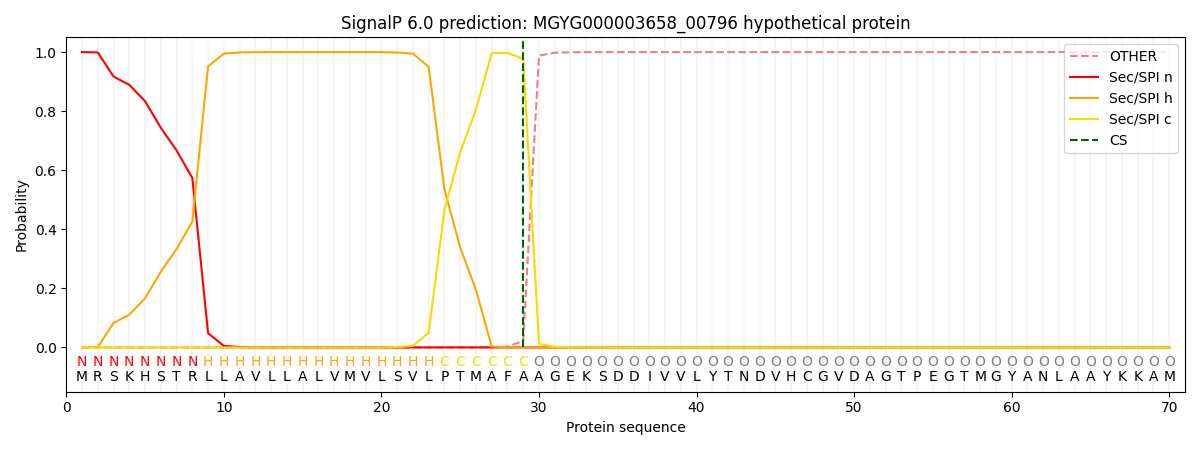
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000240 | 0.999043 | 0.000190 | 0.000182 | 0.000167 | 0.000148 |