You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003661_00928
You are here: Home > Sequence: MGYG000003661_00928
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
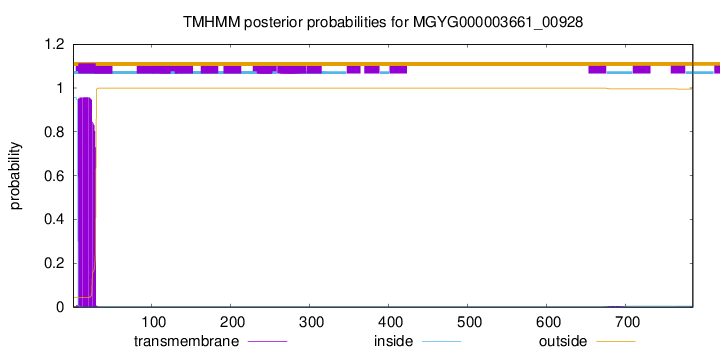
TMHMM annotations
Basic Information help
| Species | Acetatifactor sp900554205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; Acetatifactor sp900554205 | |||||||||||
| CAZyme ID | MGYG000003661_00928 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6848; End: 9205 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 109 | 455 | 2.2e-82 | 0.9834983498349835 |
| CE1 | 549 | 773 | 9e-40 | 0.8986784140969163 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 7.77e-91 | 112 | 455 | 7 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 7.85e-86 | 178 | 453 | 14 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.95e-57 | 141 | 460 | 55 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| COG2382 | Fes | 2.25e-18 | 552 | 784 | 86 | 296 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00756 | Esterase | 1.12e-12 | 551 | 698 | 11 | 142 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOZ95602.1 | 2.26e-171 | 90 | 458 | 54 | 423 |
| ADL34477.1 | 4.97e-170 | 104 | 458 | 77 | 431 |
| CBK74914.1 | 1.50e-117 | 507 | 784 | 344 | 606 |
| AKA87411.1 | 8.79e-114 | 531 | 784 | 2 | 239 |
| ADL33750.1 | 4.34e-110 | 507 | 784 | 733 | 995 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5F_A | 9.79e-56 | 99 | 457 | 176 | 528 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 1.92e-52 | 99 | 457 | 176 | 528 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2Q8X_A | 1.28e-51 | 105 | 450 | 6 | 325 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 1N82_A | 1.77e-51 | 105 | 450 | 6 | 325 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 7NL2_A | 7.99e-51 | 105 | 463 | 10 | 348 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23557 | 4.14e-71 | 153 | 452 | 1 | 297 | Putative endo-1,4-beta-xylanase OS=Caldicellulosiruptor saccharolyticus OX=44001 PE=3 SV=1 |
| P26223 | 8.79e-63 | 107 | 453 | 3 | 334 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| P51584 | 1.54e-61 | 99 | 746 | 187 | 1035 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q60042 | 2.21e-56 | 105 | 453 | 364 | 685 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q60037 | 1.42e-54 | 105 | 453 | 368 | 689 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
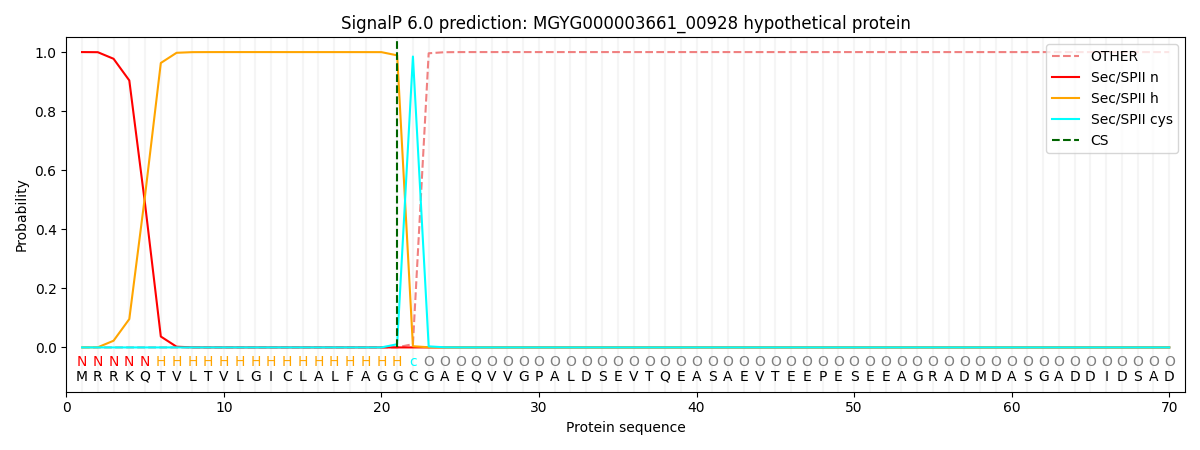
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000067 | 0.000000 | 0.000000 | 0.000000 |