You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003661_01415
You are here: Home > Sequence: MGYG000003661_01415
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acetatifactor sp900554205 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; Acetatifactor sp900554205 | |||||||||||
| CAZyme ID | MGYG000003661_01415 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12111; End: 13394 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 107 | 394 | 8.7e-100 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.10e-71 | 94 | 396 | 2 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.06e-36 | 72 | 399 | 40 | 365 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam12768 | Rax2 | 0.005 | 304 | 356 | 119 | 165 | Cortical protein marker for cell polarity. Diploid yeast cells repeatedly polarise and bud from their poles, due probably to the presence of highly stable membrane markers, and Rax2 is one such marker. It is inherited immutably at the cell cortex for multiple generations, and has a half-life exceeding several generations. The persistent inheritance of cortical protein markers would provide a means of coupling a cell's history with the future development of a precise morphogenetic form. Both Rax1 and Rax2 localize to the distal pole as well as to the division site and they interact both with each other and with Bud8p and Bud9p in the establishment and/or maintenance of the cortical markers for bipolar budding. thus Rax2 is likely to control cell polarity during vegetative growth, and in fission yeast this is done by regulating the localization of for3p. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADD61809.1 | 9.38e-164 | 61 | 427 | 68 | 438 |
| AEV59736.1 | 1.43e-163 | 57 | 427 | 72 | 440 |
| VCV21607.1 | 1.68e-163 | 61 | 427 | 85 | 455 |
| CBL14187.1 | 4.80e-163 | 61 | 427 | 85 | 455 |
| BCN29419.1 | 6.63e-159 | 61 | 427 | 77 | 448 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4X0V_A | 5.06e-152 | 58 | 426 | 22 | 395 | Structureof a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_B Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_C Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_D Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_E Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_F Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_G Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_H Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32] |
| 5H4R_A | 1.44e-151 | 58 | 426 | 22 | 395 | thecomplex of Glycoside Hydrolase 5 Lichenase from Caldicellulosiruptor sp. F32 E188Q mutant and cellotetraose [Caldicellulosiruptor sp. F32] |
| 1EDG_A | 6.74e-136 | 61 | 426 | 7 | 375 | SingleCrystal Structure Determination Of The Catalytic Domain Of Celcca Carried Out At 15 Degree C [Ruminiclostridium cellulolyticum H10] |
| 6WQP_A | 1.40e-102 | 68 | 424 | 8 | 354 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 4IM4_A | 1.46e-97 | 73 | 426 | 4 | 335 | ChainA, Endoglucanase E [Acetivibrio thermocellus],4IM4_B Chain B, Endoglucanase E [Acetivibrio thermocellus],4IM4_C Chain C, Endoglucanase E [Acetivibrio thermocellus],4IM4_D Chain D, Endoglucanase E [Acetivibrio thermocellus],4IM4_E Chain E, Endoglucanase E [Acetivibrio thermocellus],4IM4_F Chain F, Endoglucanase E [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P17901 | 8.61e-134 | 61 | 426 | 32 | 400 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P10477 | 1.44e-91 | 73 | 426 | 54 | 385 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| P20847 | 1.22e-89 | 61 | 427 | 24 | 396 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
| Q12647 | 3.98e-88 | 80 | 424 | 29 | 361 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P28623 | 4.95e-87 | 71 | 424 | 38 | 370 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
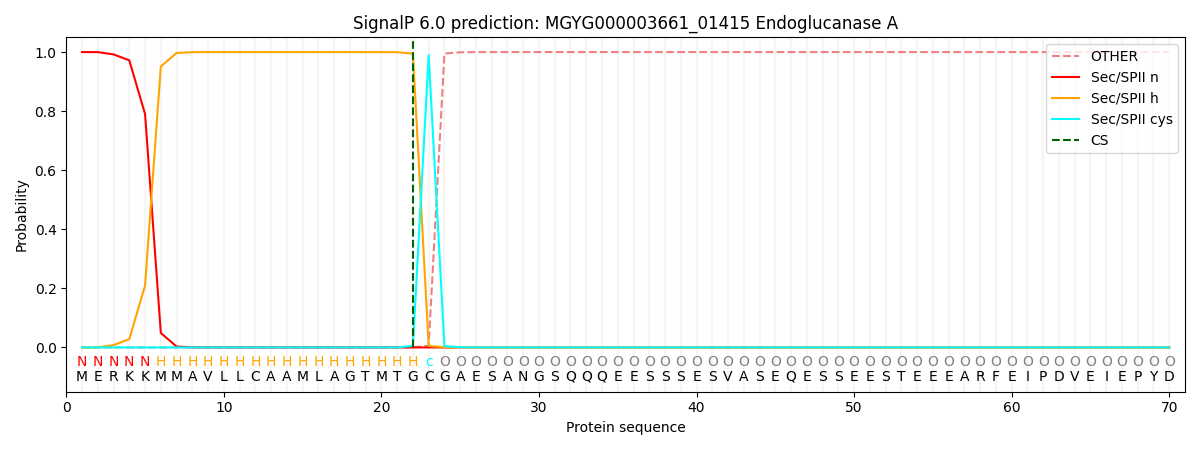
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000043 | 0.000000 | 0.000000 | 0.000000 |
