You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003662_00174
You are here: Home > Sequence: MGYG000003662_00174
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
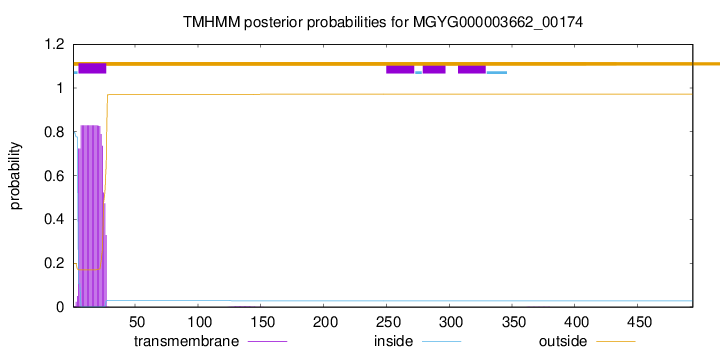
TMHMM annotations
Basic Information help
| Species | Prevotella sp900771975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900771975 | |||||||||||
| CAZyme ID | MGYG000003662_00174 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10388; End: 11872 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02671 | PLN02671 | 3.18e-13 | 196 | 401 | 61 | 272 | pectinesterase |
| PLN02773 | PLN02773 | 4.41e-12 | 198 | 400 | 9 | 213 | pectinesterase |
| PLN02176 | PLN02176 | 6.80e-10 | 187 | 407 | 32 | 253 | putative pectinesterase |
| PLN02665 | PLN02665 | 4.77e-09 | 198 | 407 | 72 | 280 | pectinesterase family protein |
| PLN02488 | PLN02488 | 1.59e-08 | 196 | 390 | 199 | 374 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65269.1 | 5.91e-309 | 1 | 494 | 38 | 531 |
| QVJ81037.1 | 1.70e-216 | 17 | 492 | 18 | 497 |
| ADE82670.1 | 6.97e-215 | 17 | 492 | 34 | 513 |
| QBI04917.1 | 2.46e-151 | 22 | 493 | 8 | 486 |
| QJD94164.1 | 2.96e-145 | 27 | 493 | 25 | 487 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FHN5 | 9.10e-07 | 175 | 351 | 201 | 387 | Probable pectinesterase/pectinesterase inhibitor 59 OS=Arabidopsis thaliana OX=3702 GN=PME59 PE=2 SV=1 |
| Q9LVQ0 | 5.23e-06 | 196 | 353 | 7 | 162 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
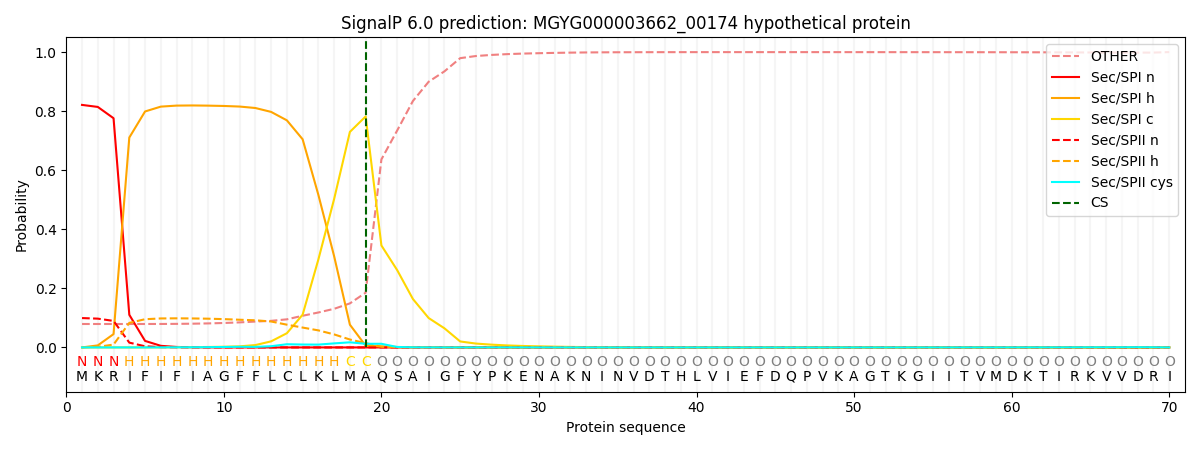
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.086015 | 0.809638 | 0.103163 | 0.000416 | 0.000324 | 0.000390 |